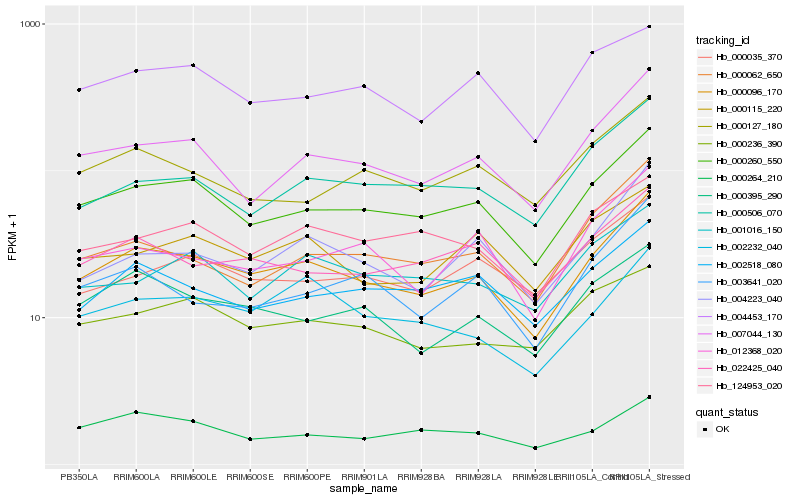
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000096_170 |
0.0 |
- |
- |
PREDICTED: 50S ribosomal protein L12, cyanelle-like [Jatropha curcas] |
| 2 |
Hb_000260_550 |
0.0615347651 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 3 |
Hb_007044_130 |
0.0853849809 |
- |
- |
PREDICTED: cyclin-B1-2 [Jatropha curcas] |
| 4 |
Hb_000395_290 |
0.0939400523 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-8 [Jatropha curcas] |
| 5 |
Hb_022425_040 |
0.096189572 |
- |
- |
PREDICTED: probable RNA methyltransferase At5g51130 [Jatropha curcas] |
| 6 |
Hb_004223_040 |
0.0976129793 |
- |
- |
PREDICTED: AP-2 complex subunit sigma [Fragaria vesca subsp. vesca] |
| 7 |
Hb_002232_040 |
0.0987910325 |
- |
- |
- |
| 8 |
Hb_002518_080 |
0.0995214668 |
- |
- |
PREDICTED: uncharacterized protein LOC105639837 [Jatropha curcas] |
| 9 |
Hb_124953_020 |
0.1017525384 |
- |
- |
Mitochondrial import inner membrane translocase subunit Tim13, putative [Ricinus communis] |
| 10 |
Hb_000506_070 |
0.1034410057 |
- |
- |
PREDICTED: 60S ribosomal protein L35 [Jatropha curcas] |
| 11 |
Hb_004453_170 |
0.1043434035 |
- |
- |
hypothetical protein POPTR_0018s01370g [Populus trichocarpa] |
| 12 |
Hb_003641_020 |
0.1050582493 |
- |
- |
hypothetical protein POPTR_0006s13960g [Populus trichocarpa] |
| 13 |
Hb_000115_220 |
0.1067844597 |
- |
- |
hypothetical protein CISIN_1g0379742mg, partial [Citrus sinensis] |
| 14 |
Hb_000062_650 |
0.1069501019 |
- |
- |
PREDICTED: uncharacterized protein LOC105628245 [Jatropha curcas] |
| 15 |
Hb_000264_210 |
0.1082185142 |
- |
- |
tfiih, polypeptide, putative [Ricinus communis] |
| 16 |
Hb_000236_390 |
0.1090129023 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH1 [Jatropha curcas] |
| 17 |
Hb_001016_150 |
0.1093830533 |
- |
- |
Rop1 [Hevea brasiliensis] |
| 18 |
Hb_012368_020 |
0.1094246204 |
- |
- |
PREDICTED: uncharacterized protein LOC105650964 [Jatropha curcas] |
| 19 |
Hb_000127_180 |
0.1096927829 |
- |
- |
40S ribosomal protein S18, putative [Ricinus communis] |
| 20 |
Hb_000035_370 |
0.1098291342 |
- |
- |
conserved hypothetical protein [Ricinus communis] |