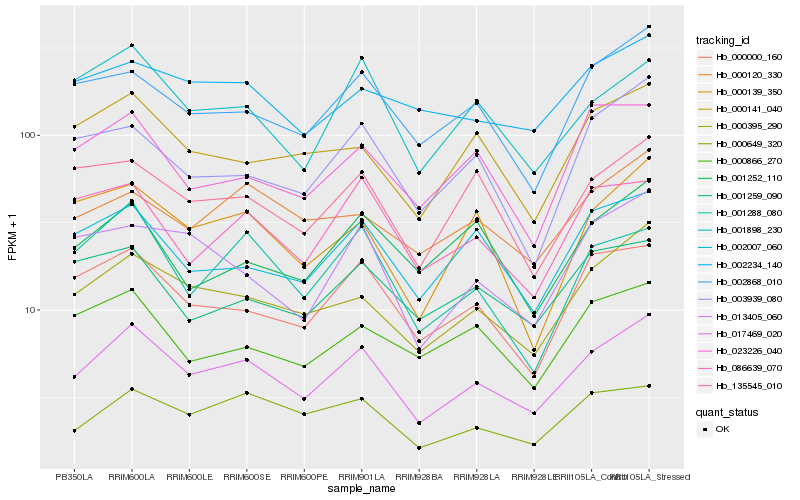
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017469_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000395_290 |
0.0802222304 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-8 [Jatropha curcas] |
| 3 |
Hb_000000_160 |
0.0960136033 |
- |
- |
PREDICTED: membrin-11 [Jatropha curcas] |
| 4 |
Hb_002868_010 |
0.098926621 |
- |
- |
60S ribosomal protein L30, putative [Ricinus communis] |
| 5 |
Hb_000649_320 |
0.1003023626 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002007_060 |
0.1021340174 |
- |
- |
hypothetical protein JCGZ_23323 [Jatropha curcas] |
| 7 |
Hb_001898_230 |
0.1045844665 |
- |
- |
PREDICTED: KH domain-containing protein At1g09660/At1g09670 isoform X2 [Jatropha curcas] |
| 8 |
Hb_135545_010 |
0.1081573396 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 9 |
Hb_000141_040 |
0.1083383819 |
- |
- |
hypothetical protein CICLE_v10022858mg [Citrus clementina] |
| 10 |
Hb_000120_330 |
0.1104952303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001288_080 |
0.1109824994 |
- |
- |
PREDICTED: probable protein phosphatase 2C 50 [Populus euphratica] |
| 12 |
Hb_086639_070 |
0.1112619681 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM2 [Jatropha curcas] |
| 13 |
Hb_013405_060 |
0.1112992927 |
- |
- |
PREDICTED: probable RNA-binding protein 18 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000139_350 |
0.1116685995 |
- |
- |
AtGCP3 interacting protein 1 [Theobroma cacao] |
| 15 |
Hb_002234_140 |
0.1124935347 |
- |
- |
DNA-directed RNA polymerase II family protein [Populus trichocarpa] |
| 16 |
Hb_003939_080 |
0.1140868185 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000866_270 |
0.1146764009 |
- |
- |
PREDICTED: uncharacterized protein LOC105642993 [Jatropha curcas] |
| 18 |
Hb_023226_040 |
0.1163451329 |
- |
- |
PREDICTED: kxDL motif-containing protein 1 [Jatropha curcas] |
| 19 |
Hb_001259_090 |
0.1164039701 |
- |
- |
hypothetical protein B456_005G056700 [Gossypium raimondii] |
| 20 |
Hb_001252_110 |
0.1180773423 |
- |
- |
PREDICTED: uncharacterized protein At2g39795, mitochondrial-like [Jatropha curcas] |