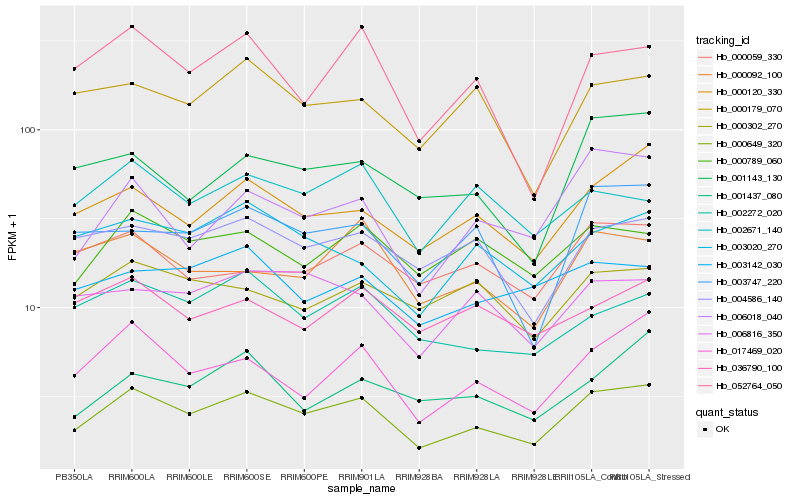
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000649_320 |
0.0 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000120_330 |
0.096622269 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_017469_020 |
0.1003023626 |
- |
- |
- |
| 4 |
Hb_000789_060 |
0.1072380938 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 6 [Jatropha curcas] |
| 5 |
Hb_006018_040 |
0.1077921787 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 6 |
Hb_004586_140 |
0.1096475876 |
- |
- |
PREDICTED: uncharacterized protein LOC105630689 [Jatropha curcas] |
| 7 |
Hb_052764_050 |
0.1122707002 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 8 |
Hb_002272_020 |
0.1129830038 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 9 |
Hb_003020_270 |
0.1165522685 |
- |
- |
PREDICTED: ras-related protein Rab2BV [Jatropha curcas] |
| 10 |
Hb_001143_130 |
0.1174460508 |
- |
- |
peroxidase 2 [Hevea brasiliensis] |
| 11 |
Hb_003747_220 |
0.1178013019 |
- |
- |
PREDICTED: syntaxin-22-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_000179_070 |
0.118293953 |
- |
- |
hypothetical protein CICLE_v10009411mg [Citrus clementina] |
| 13 |
Hb_006816_350 |
0.1185129277 |
- |
- |
PREDICTED: syntaxin-43 [Jatropha curcas] |
| 14 |
Hb_000092_100 |
0.1196377509 |
- |
- |
PREDICTED: protein EMSY-LIKE 1 [Jatropha curcas] |
| 15 |
Hb_000302_270 |
0.1210628967 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_000059_330 |
0.121469125 |
- |
- |
PREDICTED: polyadenylate-binding protein 1 [Jatropha curcas] |
| 17 |
Hb_002671_140 |
0.122244492 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 18 |
Hb_001437_080 |
0.1222626776 |
transcription factor |
TF Family: TCP |
hypothetical protein JCGZ_05472 [Jatropha curcas] |
| 19 |
Hb_003142_030 |
0.1224524001 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 14, chloroplastic [Jatropha curcas] |
| 20 |
Hb_036790_100 |
0.1236093195 |
- |
- |
PREDICTED: uncharacterized protein LOC105645120 [Jatropha curcas] |