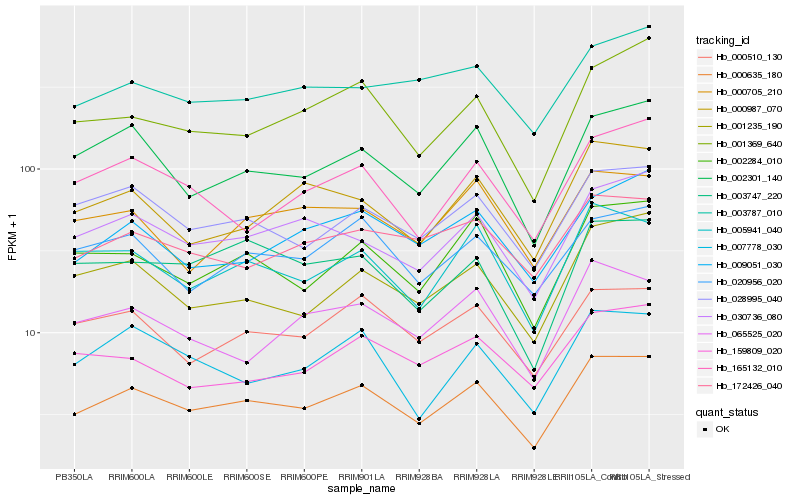
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000635_180 |
0.0 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002284_010 |
0.0744624878 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002301_140 |
0.0759069179 |
- |
- |
PREDICTED: uncharacterized protein At4g28440 [Jatropha curcas] |
| 4 |
Hb_028995_040 |
0.0769144988 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 11 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005941_040 |
0.0782492031 |
- |
- |
PREDICTED: CASP-like protein 5A2 [Jatropha curcas] |
| 6 |
Hb_030736_080 |
0.078830879 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001235_190 |
0.0822535123 |
- |
- |
PREDICTED: peroxisome biogenesis protein 19-2-like [Jatropha curcas] |
| 8 |
Hb_000510_130 |
0.082622905 |
- |
- |
PREDICTED: PRA1 family protein F2-like [Jatropha curcas] |
| 9 |
Hb_020956_020 |
0.087665195 |
- |
- |
PREDICTED: SNF1-related protein kinase regulatory subunit beta-3-like [Jatropha curcas] |
| 10 |
Hb_001369_640 |
0.0886064984 |
- |
- |
PREDICTED: 40S ribosomal protein S20-1 [Jatropha curcas] |
| 11 |
Hb_009051_030 |
0.0889454076 |
- |
- |
PREDICTED: thioredoxin O1, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_172426_040 |
0.089602611 |
- |
- |
PREDICTED: ELL-associated factor 1 [Jatropha curcas] |
| 13 |
Hb_000987_070 |
0.0901591101 |
- |
- |
Inflorescence meristem receptor-like kinase 2 isoform 1 [Theobroma cacao] |
| 14 |
Hb_007778_030 |
0.0913440584 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_065525_020 |
0.0917463383 |
- |
- |
PREDICTED: geranylgeranyl transferase type-1 subunit beta [Jatropha curcas] |
| 16 |
Hb_000705_210 |
0.0920791457 |
- |
- |
PREDICTED: ras-related protein Rab2BV [Jatropha curcas] |
| 17 |
Hb_003747_220 |
0.0925331838 |
- |
- |
PREDICTED: syntaxin-22-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_003787_010 |
0.093340276 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3-like [Phoenix dactylifera] |
| 19 |
Hb_165132_010 |
0.0936952637 |
- |
- |
PREDICTED: uncharacterized protein LOC105134496 [Populus euphratica] |
| 20 |
Hb_159809_020 |
0.0950396573 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |