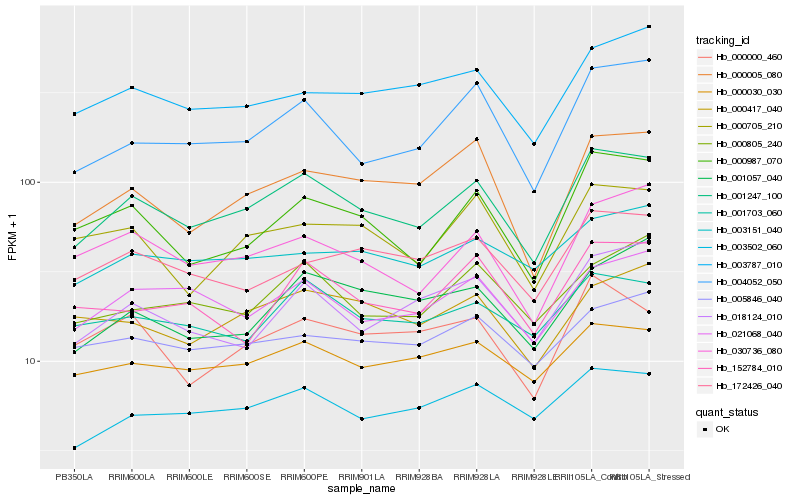
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001247_100 |
0.0 |
- |
- |
PREDICTED: pyroglutamyl-peptidase 1-like protein [Jatropha curcas] |
| 2 |
Hb_000987_070 |
0.0805054894 |
- |
- |
Inflorescence meristem receptor-like kinase 2 isoform 1 [Theobroma cacao] |
| 3 |
Hb_003502_060 |
0.0834557356 |
- |
- |
PREDICTED: alpha-1,3/1,6-mannosyltransferase ALG2 [Jatropha curcas] |
| 4 |
Hb_004052_050 |
0.0836453631 |
- |
- |
PREDICTED: uncharacterized protein LOC105640640 [Jatropha curcas] |
| 5 |
Hb_000005_080 |
0.0845707914 |
- |
- |
PREDICTED: cyclin-dependent kinases regulatory subunit 1 [Elaeis guineensis] |
| 6 |
Hb_021068_040 |
0.0850031534 |
- |
- |
PREDICTED: XIAP-associated factor 1 [Jatropha curcas] |
| 7 |
Hb_030736_080 |
0.0851018603 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000030_030 |
0.0879827897 |
- |
- |
PREDICTED: ADP-glucose phosphorylase [Jatropha curcas] |
| 9 |
Hb_001703_060 |
0.0887876438 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 10 |
Hb_000705_210 |
0.0900094448 |
- |
- |
PREDICTED: ras-related protein Rab2BV [Jatropha curcas] |
| 11 |
Hb_000417_040 |
0.0924522279 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 12 |
Hb_172426_040 |
0.0925607935 |
- |
- |
PREDICTED: ELL-associated factor 1 [Jatropha curcas] |
| 13 |
Hb_003787_010 |
0.0927743157 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3-like [Phoenix dactylifera] |
| 14 |
Hb_000805_240 |
0.0930106155 |
- |
- |
PREDICTED: uncharacterized protein C20orf24 homolog [Jatropha curcas] |
| 15 |
Hb_018124_010 |
0.0938808004 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000000_460 |
0.0940144667 |
- |
- |
PREDICTED: peroxisomal membrane protein 11A [Jatropha curcas] |
| 17 |
Hb_152784_010 |
0.0950482713 |
- |
- |
hypothetical protein PRUPE_ppa010913mg [Prunus persica] |
| 18 |
Hb_003151_040 |
0.0954695535 |
transcription factor |
TF Family: Alfin-like |
zinc finger family protein [Populus trichocarpa] |
| 19 |
Hb_005846_040 |
0.0959640998 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 4 [Populus euphratica] |
| 20 |
Hb_001057_040 |
0.0966860889 |
- |
- |
PREDICTED: oligosaccharyltransferase complex subunit ostc [Jatropha curcas] |