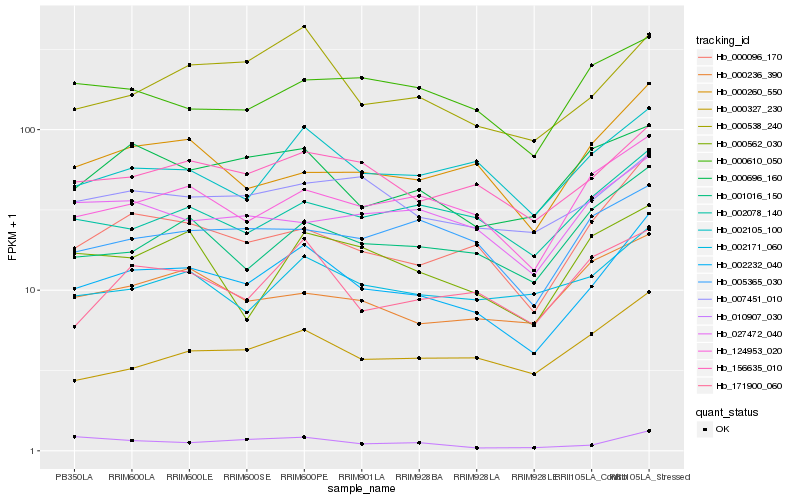
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002232_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_000096_170 |
0.0987910325 |
- |
- |
PREDICTED: 50S ribosomal protein L12, cyanelle-like [Jatropha curcas] |
| 3 |
Hb_156635_010 |
0.0990747651 |
- |
- |
PREDICTED: autophagy-related protein 8f [Jatropha curcas] |
| 4 |
Hb_124953_020 |
0.1072805895 |
- |
- |
Mitochondrial import inner membrane translocase subunit Tim13, putative [Ricinus communis] |
| 5 |
Hb_000538_240 |
0.1078887881 |
- |
- |
PREDICTED: uncharacterized protein LOC105643073 [Jatropha curcas] |
| 6 |
Hb_002105_100 |
0.1100754265 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_001016_150 |
0.1187089394 |
- |
- |
Rop1 [Hevea brasiliensis] |
| 8 |
Hb_002171_060 |
0.1194833915 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007451_010 |
0.1197043361 |
- |
- |
PREDICTED: 60S ribosome subunit biogenesis protein NIP7 homolog [Jatropha curcas] |
| 10 |
Hb_002078_140 |
0.1205116951 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 11 |
Hb_171900_060 |
0.1208388294 |
- |
- |
PREDICTED: uncharacterized protein LOC105643368 [Jatropha curcas] |
| 12 |
Hb_000327_230 |
0.1244457405 |
- |
- |
PREDICTED: nitrilase-like protein 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000696_160 |
0.1245419393 |
- |
- |
PREDICTED: SKP1-like protein 1B [Jatropha curcas] |
| 14 |
Hb_010907_030 |
0.1262233584 |
- |
- |
- |
| 15 |
Hb_000260_550 |
0.126679846 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 16 |
Hb_005365_030 |
0.127584158 |
- |
- |
hypothetical protein POPTR_0002s24010g [Populus trichocarpa] |
| 17 |
Hb_000562_030 |
0.1288883295 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_027472_040 |
0.1305700654 |
- |
- |
SKP1 [Hevea brasiliensis] |
| 19 |
Hb_000610_050 |
0.132210649 |
- |
- |
skp1, putative [Ricinus communis] |
| 20 |
Hb_000236_390 |
0.1331404669 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH1 [Jatropha curcas] |