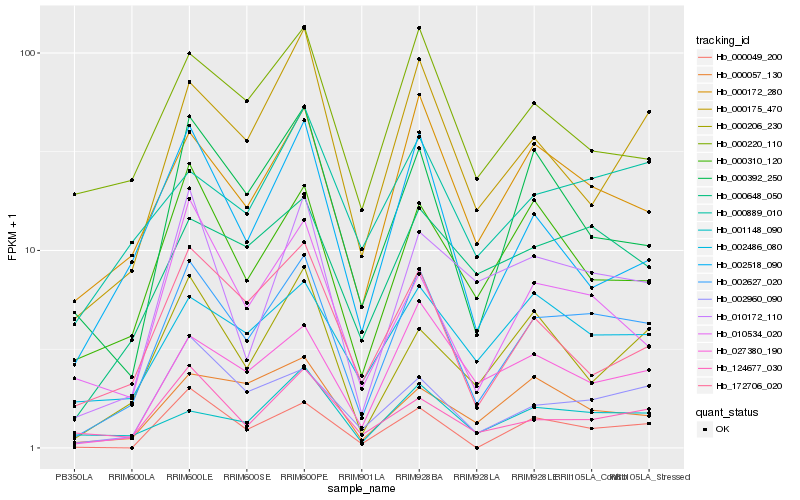
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002627_020 |
0.0 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 2 |
Hb_002960_090 |
0.1386565764 |
- |
- |
protein with unknown function [Ricinus communis] |
| 3 |
Hb_000172_280 |
0.1393968744 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 4 |
Hb_000175_470 |
0.149148408 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 5 |
Hb_027380_190 |
0.1552568221 |
- |
- |
PREDICTED: uncharacterized protein LOC105634028 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000310_120 |
0.1560091026 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 7 |
Hb_000648_050 |
0.1577870592 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000392_250 |
0.1578425751 |
- |
- |
PREDICTED: HVA22-like protein a [Jatropha curcas] |
| 9 |
Hb_010534_020 |
0.1590257913 |
- |
- |
PREDICTED: protein XRI1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000049_200 |
0.1595104142 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 11 |
Hb_124677_030 |
0.1605399001 |
- |
- |
PREDICTED: uncharacterized protein LOC105650473 [Jatropha curcas] |
| 12 |
Hb_000206_230 |
0.1623759187 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_172706_020 |
0.1625726383 |
- |
- |
PREDICTED: protein ABIL2 [Jatropha curcas] |
| 14 |
Hb_002518_090 |
0.163643036 |
- |
- |
PREDICTED: tubulin beta-2 chain [Jatropha curcas] |
| 15 |
Hb_010172_110 |
0.1643444926 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 16 |
Hb_001148_090 |
0.1684866257 |
- |
- |
hypothetical protein JCGZ_14711 [Jatropha curcas] |
| 17 |
Hb_002486_080 |
0.1691431437 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 18 |
Hb_000057_130 |
0.1718776406 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 19 |
Hb_000220_110 |
0.1731175022 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 20 |
Hb_000889_010 |
0.1743167264 |
transcription factor |
TF Family: MYB-related |
hypothetical protein PRUPE_ppa014728mg [Prunus persica] |