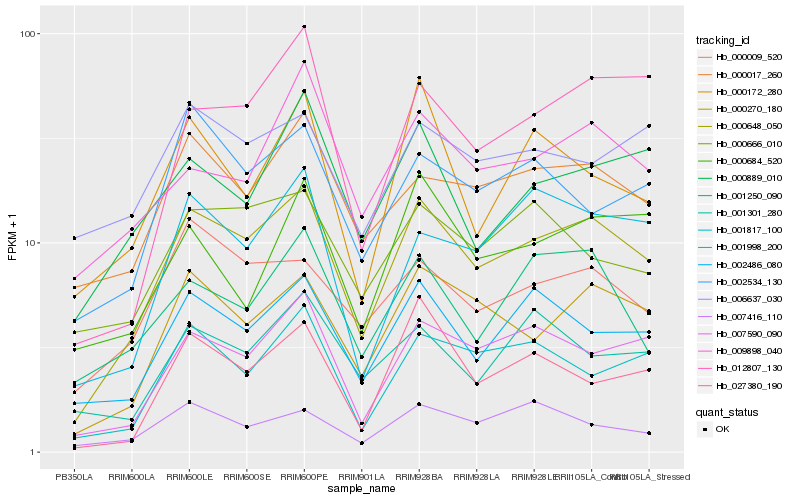
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000648_050 |
0.0 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000270_180 |
0.103612774 |
- |
- |
PREDICTED: HIPL1 protein-like [Jatropha curcas] |
| 3 |
Hb_007590_090 |
0.1156324923 |
- |
- |
PREDICTED: O-glucosyltransferase rumi [Jatropha curcas] |
| 4 |
Hb_002486_080 |
0.1194293117 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 5 |
Hb_000017_260 |
0.1285443399 |
- |
- |
hypothetical protein POPTR_0003s13070g [Populus trichocarpa] |
| 6 |
Hb_001817_100 |
0.1346475584 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 7 |
Hb_000009_520 |
0.1356959827 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 14 [Jatropha curcas] |
| 8 |
Hb_012807_130 |
0.1376541436 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 9 |
Hb_001998_200 |
0.1384619337 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 10 |
Hb_002534_130 |
0.1434986473 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 11 |
Hb_000889_010 |
0.1435441682 |
transcription factor |
TF Family: MYB-related |
hypothetical protein PRUPE_ppa014728mg [Prunus persica] |
| 12 |
Hb_009898_040 |
0.1438728929 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000666_010 |
0.1444960527 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001250_090 |
0.1452989482 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 3 [Jatropha curcas] |
| 15 |
Hb_007416_110 |
0.1470534248 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 16 |
Hb_027380_190 |
0.1492970701 |
- |
- |
PREDICTED: uncharacterized protein LOC105634028 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000684_520 |
0.1495176662 |
- |
- |
glutathione S-transferase L3-like [Jatropha curcas] |
| 18 |
Hb_001301_280 |
0.150940038 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000172_280 |
0.1526276661 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 20 |
Hb_006637_030 |
0.1531601869 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |