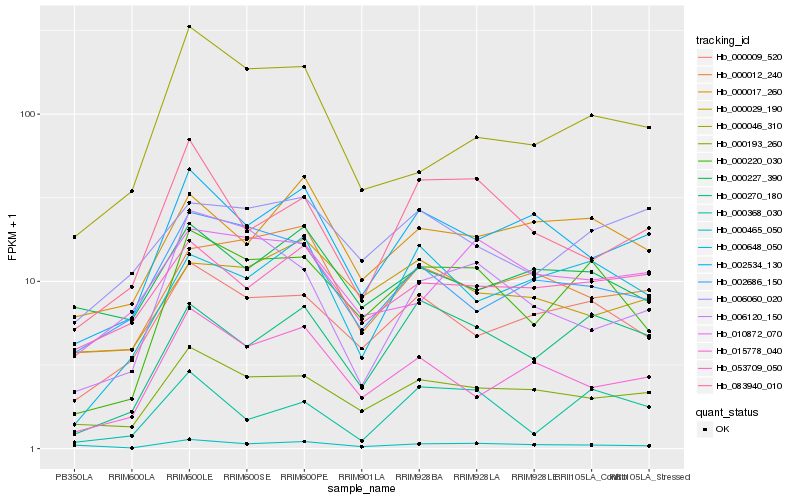
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000220_030 |
0.0 |
- |
- |
PREDICTED: amino-acid permease BAT1 homolog isoform X2 [Jatropha curcas] |
| 2 |
Hb_000270_180 |
0.1434499457 |
- |
- |
PREDICTED: HIPL1 protein-like [Jatropha curcas] |
| 3 |
Hb_000009_520 |
0.1456834383 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 14 [Jatropha curcas] |
| 4 |
Hb_000193_260 |
0.1578187872 |
- |
- |
PREDICTED: protein NEN1 [Jatropha curcas] |
| 5 |
Hb_000648_050 |
0.1757933388 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000017_260 |
0.183983333 |
- |
- |
hypothetical protein POPTR_0003s13070g [Populus trichocarpa] |
| 7 |
Hb_002534_130 |
0.1880792127 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 8 |
Hb_000368_030 |
0.1901381007 |
- |
- |
PREDICTED: cytokinin dehydrogenase 5 [Jatropha curcas] |
| 9 |
Hb_000465_050 |
0.1904025844 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH4 [Jatropha curcas] |
| 10 |
Hb_002686_150 |
0.1905345619 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 11 |
Hb_000012_240 |
0.1955546503 |
- |
- |
PREDICTED: ferrochelatase-2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_010872_070 |
0.1957178661 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_015778_040 |
0.1972744017 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |
| 14 |
Hb_006120_150 |
0.1979477177 |
- |
- |
invertase [Hevea brasiliensis] |
| 15 |
Hb_053709_050 |
0.1993216849 |
- |
- |
PREDICTED: nudix hydrolase 23, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_000029_190 |
0.1994222404 |
- |
- |
hypothetical protein CISIN_1g025338mg [Citrus sinensis] |
| 17 |
Hb_006060_020 |
0.1995769047 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 8 isoform X3 [Jatropha curcas] |
| 18 |
Hb_083940_010 |
0.2019399401 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 19 |
Hb_000227_390 |
0.2022993902 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000046_310 |
0.2055973717 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |