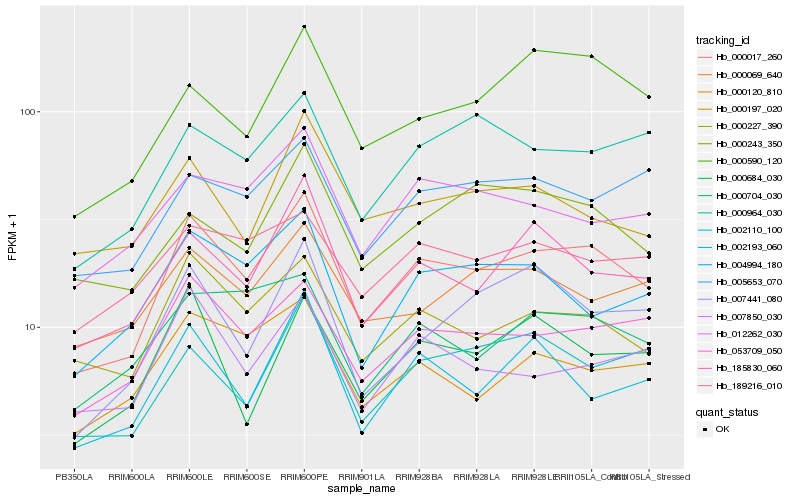
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000017_260 |
0.0 |
- |
- |
hypothetical protein POPTR_0003s13070g [Populus trichocarpa] |
| 2 |
Hb_053709_050 |
0.0872785313 |
- |
- |
PREDICTED: nudix hydrolase 23, chloroplastic isoform X2 [Jatropha curcas] |
| 3 |
Hb_000227_390 |
0.0922679547 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_185830_060 |
0.0924545486 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 5 |
Hb_002193_060 |
0.10565457 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 6 |
Hb_012262_030 |
0.1080052071 |
- |
- |
PREDICTED: thylakoidal processing peptidase 1, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_000590_120 |
0.1080439646 |
- |
- |
dehydroascorbate reductase, putative [Ricinus communis] |
| 8 |
Hb_004994_180 |
0.1083396859 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 9 |
Hb_002110_100 |
0.1084714732 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000964_030 |
0.1086755801 |
- |
- |
ADP/ATP carrier 2 [Theobroma cacao] |
| 11 |
Hb_007850_030 |
0.109015307 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 12 |
Hb_005653_070 |
0.1094633154 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 13 |
Hb_000120_810 |
0.1106309967 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 14 |
Hb_007441_080 |
0.1116862145 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 15 |
Hb_000684_030 |
0.1129831099 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 16 |
Hb_000704_030 |
0.1135234716 |
- |
- |
PREDICTED: probable aquaporin SIP2-1 [Cicer arietinum] |
| 17 |
Hb_000069_640 |
0.1140623445 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 18 |
Hb_189216_010 |
0.1150653175 |
- |
- |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000197_020 |
0.1152235276 |
- |
- |
PREDICTED: protein RER1A [Jatropha curcas] |
| 20 |
Hb_000243_350 |
0.1157821508 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Tarenaya hassleriana] |