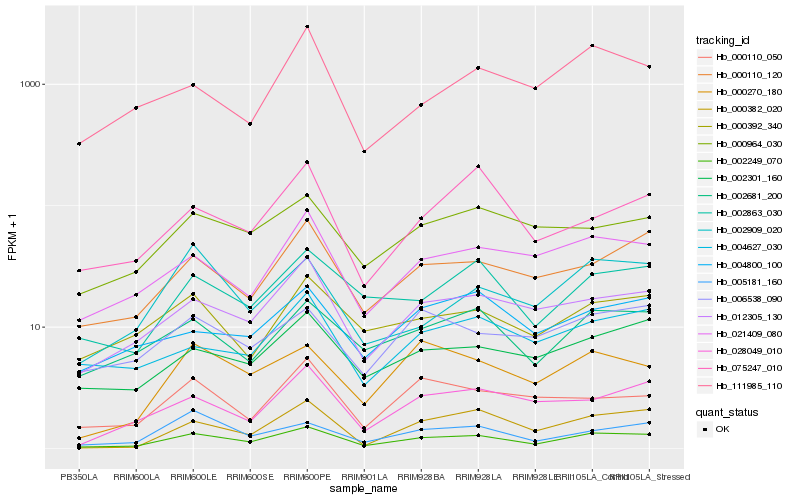
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002249_070 |
0.0 |
- |
- |
gcn4-complementing protein, putative [Ricinus communis] |
| 2 |
Hb_012305_130 |
0.1257187425 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 3 |
Hb_000110_120 |
0.1295378111 |
- |
- |
PREDICTED: ras-related protein RABE1a [Fragaria vesca subsp. vesca] |
| 4 |
Hb_000270_180 |
0.1383322796 |
- |
- |
PREDICTED: HIPL1 protein-like [Jatropha curcas] |
| 5 |
Hb_021409_080 |
0.1385750385 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 6 |
Hb_000382_020 |
0.1388509103 |
- |
- |
PREDICTED: uncharacterized protein LOC105633610 [Jatropha curcas] |
| 7 |
Hb_002863_030 |
0.1389281614 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 8 |
Hb_002681_200 |
0.1394621193 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 [Jatropha curcas] |
| 9 |
Hb_002301_160 |
0.142748119 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04780-like [Jatropha curcas] |
| 10 |
Hb_005181_160 |
0.1428545556 |
- |
- |
PREDICTED: uncharacterized protein LOC105643546 [Jatropha curcas] |
| 11 |
Hb_028049_010 |
0.1431844233 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002909_020 |
0.1520146901 |
- |
- |
PREDICTED: uncharacterized protein LOC105632476 [Jatropha curcas] |
| 13 |
Hb_000392_340 |
0.1553359644 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 29 [Jatropha curcas] |
| 14 |
Hb_000110_050 |
0.1564096055 |
- |
- |
- |
| 15 |
Hb_000964_030 |
0.157048073 |
- |
- |
ADP/ATP carrier 2 [Theobroma cacao] |
| 16 |
Hb_075247_010 |
0.1579443136 |
- |
- |
PREDICTED: actin-depolymerizing factor 2-like [Nelumbo nucifera] |
| 17 |
Hb_006538_090 |
0.159132438 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 18 |
Hb_004627_030 |
0.1595131044 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 19 |
Hb_111985_110 |
0.1601809066 |
- |
- |
thioredoxin H-type 3 [Hevea brasiliensis] |
| 20 |
Hb_004800_100 |
0.1617655583 |
- |
- |
conserved hypothetical protein [Ricinus communis] |