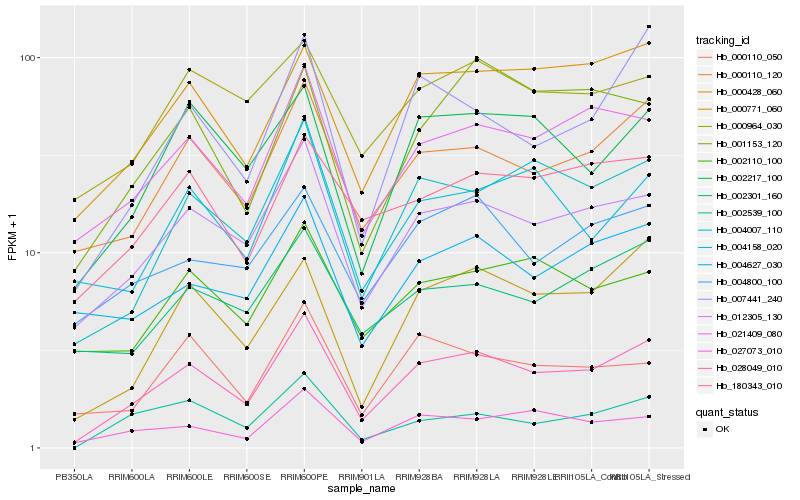
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028049_010 |
0.0 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_012305_130 |
0.0919426333 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 3 |
Hb_000110_120 |
0.1038496645 |
- |
- |
PREDICTED: ras-related protein RABE1a [Fragaria vesca subsp. vesca] |
| 4 |
Hb_004007_110 |
0.1076917042 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 5 |
Hb_021409_080 |
0.1132193341 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 6 |
Hb_000771_060 |
0.1156775531 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |
| 7 |
Hb_000428_060 |
0.116466483 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_027073_010 |
0.121913 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 9 |
Hb_007441_240 |
0.1239335025 |
- |
- |
ethphon-induced protein [Hevea brasiliensis] |
| 10 |
Hb_002217_100 |
0.1246382916 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
dihydrolipoamide dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_002539_100 |
0.1332326536 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 iota [Jatropha curcas] |
| 12 |
Hb_180343_010 |
0.1361828858 |
- |
- |
Biotin carboxyl carrier protein of acetyl-CoA carboxylase, putative [Ricinus communis] |
| 13 |
Hb_004800_100 |
0.1380129653 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002110_100 |
0.1394330506 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002301_160 |
0.1401395322 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04780-like [Jatropha curcas] |
| 16 |
Hb_000110_050 |
0.1403239499 |
- |
- |
- |
| 17 |
Hb_004158_020 |
0.1404123902 |
- |
- |
Rab1 [Hevea brasiliensis] |
| 18 |
Hb_004627_030 |
0.1411678277 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 19 |
Hb_001153_120 |
0.1423869599 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 20 |
Hb_000964_030 |
0.1425570161 |
- |
- |
ADP/ATP carrier 2 [Theobroma cacao] |