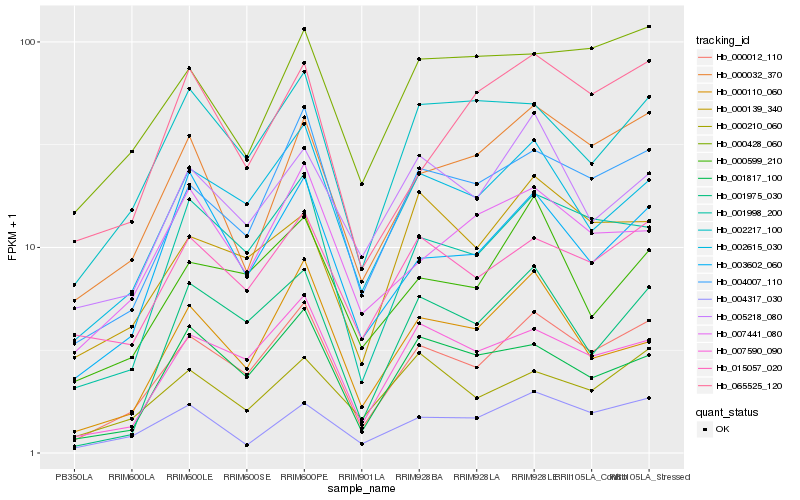
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000012_110 |
0.0 |
- |
- |
hypothetical protein CICLE_v10008613mg [Citrus clementina] |
| 2 |
Hb_004007_110 |
0.0829108833 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 3 |
Hb_001998_200 |
0.086182866 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 4 |
Hb_002615_030 |
0.0975577884 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 5 |
Hb_007590_090 |
0.0976473162 |
- |
- |
PREDICTED: O-glucosyltransferase rumi [Jatropha curcas] |
| 6 |
Hb_003602_060 |
0.1153245197 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein 1 [Jatropha curcas] |
| 7 |
Hb_015057_020 |
0.1157467545 |
- |
- |
PREDICTED: E3 SUMO-protein ligase MMS21 [Jatropha curcas] |
| 8 |
Hb_000032_370 |
0.1164784318 |
- |
- |
PREDICTED: nucleoside diphosphate kinase III, chloroplastic/mitochondrial-like [Citrus sinensis] |
| 9 |
Hb_001975_030 |
0.1198176311 |
- |
- |
4-diphosphocytidyl-2-c-methyl-d-erythritol kinase, partial [Plectranthus barbatus] |
| 10 |
Hb_002217_100 |
0.1206708343 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
dihydrolipoamide dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_001817_100 |
0.1222274272 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 12 |
Hb_000599_210 |
0.1277717341 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 13 |
Hb_004317_030 |
0.1280150947 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 14 |
Hb_000139_340 |
0.1284214468 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000210_060 |
0.1320989825 |
transcription factor |
TF Family: E2F-DP |
hypothetical protein JCGZ_08780 [Jatropha curcas] |
| 16 |
Hb_000110_060 |
0.1328074852 |
- |
- |
PREDICTED: uncharacterized protein LOC105636258 [Jatropha curcas] |
| 17 |
Hb_000428_060 |
0.1341499148 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_005218_080 |
0.134506915 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_065525_120 |
0.1347439218 |
- |
- |
dtdp-glucose 4-6-dehydratase, putative [Ricinus communis] |
| 20 |
Hb_007441_080 |
0.1379102447 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |