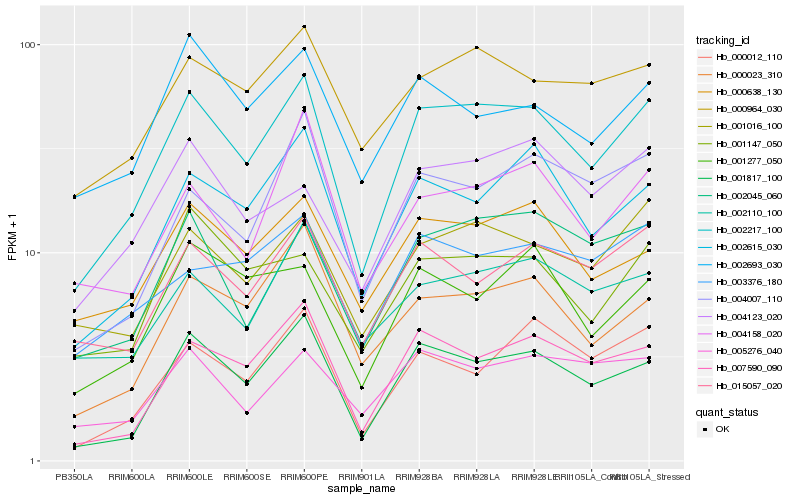
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002217_100 |
0.0 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
dihydrolipoamide dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_001817_100 |
0.0873966543 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 3 |
Hb_002615_030 |
0.0996307308 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 4 |
Hb_001016_100 |
0.1026841092 |
- |
- |
PREDICTED: V-type proton ATPase subunit E-like [Pyrus x bretschneideri] |
| 5 |
Hb_007590_090 |
0.1043204796 |
- |
- |
PREDICTED: O-glucosyltransferase rumi [Jatropha curcas] |
| 6 |
Hb_002045_060 |
0.1081476042 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000964_030 |
0.1101182978 |
- |
- |
ADP/ATP carrier 2 [Theobroma cacao] |
| 8 |
Hb_000638_130 |
0.1103140156 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 9 |
Hb_015057_020 |
0.1130797019 |
- |
- |
PREDICTED: E3 SUMO-protein ligase MMS21 [Jatropha curcas] |
| 10 |
Hb_000023_310 |
0.1155026286 |
- |
- |
PREDICTED: probable folate-biopterin transporter 2 [Populus euphratica] |
| 11 |
Hb_002110_100 |
0.1170413919 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_004123_020 |
0.1198540425 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_000012_110 |
0.1206708343 |
- |
- |
hypothetical protein CICLE_v10008613mg [Citrus clementina] |
| 14 |
Hb_005276_040 |
0.1215337603 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2-like [Jatropha curcas] |
| 15 |
Hb_001147_050 |
0.1220279676 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase Cx32, chloroplastic [Jatropha curcas] |
| 16 |
Hb_003376_180 |
0.1221061302 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 17 |
Hb_004007_110 |
0.1226360966 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 18 |
Hb_004158_020 |
0.1238468725 |
- |
- |
Rab1 [Hevea brasiliensis] |
| 19 |
Hb_001277_050 |
0.1239097233 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002693_030 |
0.123941383 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 34 kDa [Jatropha curcas] |