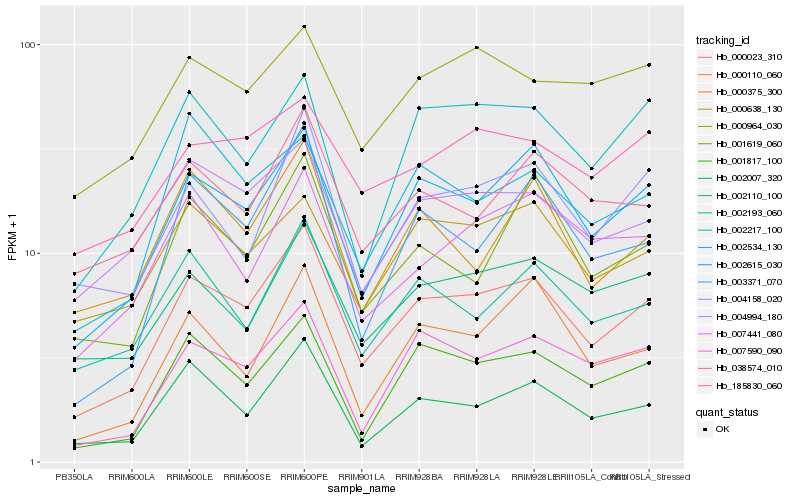
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000023_310 |
0.0 |
- |
- |
PREDICTED: probable folate-biopterin transporter 2 [Populus euphratica] |
| 2 |
Hb_002615_030 |
0.0754788555 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 3 |
Hb_002007_320 |
0.1039062789 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase ANP1 [Jatropha curcas] |
| 4 |
Hb_001817_100 |
0.1064512622 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 5 |
Hb_004158_020 |
0.1075483636 |
- |
- |
Rab1 [Hevea brasiliensis] |
| 6 |
Hb_007590_090 |
0.108511426 |
- |
- |
PREDICTED: O-glucosyltransferase rumi [Jatropha curcas] |
| 7 |
Hb_003371_070 |
0.1088346792 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 8 |
Hb_004994_180 |
0.1092787594 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 9 |
Hb_002193_060 |
0.1114871843 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 10 |
Hb_002110_100 |
0.1136476624 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002217_100 |
0.1155026286 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
dihydrolipoamide dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_185830_060 |
0.1186129332 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 13 |
Hb_000110_060 |
0.1195330346 |
- |
- |
PREDICTED: uncharacterized protein LOC105636258 [Jatropha curcas] |
| 14 |
Hb_007441_080 |
0.1203838979 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 15 |
Hb_001619_060 |
0.1206965007 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000638_130 |
0.1209958012 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 17 |
Hb_000375_300 |
0.1214654763 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 18 |
Hb_038574_010 |
0.125529401 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 3-like isoform X2 [Populus euphratica] |
| 19 |
Hb_000964_030 |
0.1269968575 |
- |
- |
ADP/ATP carrier 2 [Theobroma cacao] |
| 20 |
Hb_002534_130 |
0.1272200641 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |