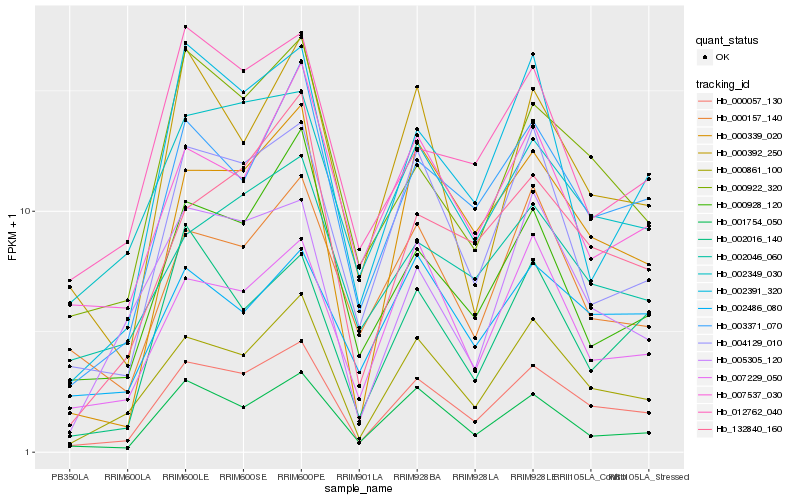
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000057_130 |
0.0 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 2 |
Hb_000861_100 |
0.0795967883 |
- |
- |
PREDICTED: probable fucosyltransferase 8 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003371_070 |
0.102267965 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 4 |
Hb_001754_050 |
0.1083030893 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 5 |
Hb_004129_010 |
0.1094747444 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 6 |
Hb_000339_020 |
0.1128706069 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 12-like [Jatropha curcas] |
| 7 |
Hb_000922_320 |
0.11654983 |
- |
- |
hypothetical protein B456_001G071400 [Gossypium raimondii] |
| 8 |
Hb_002349_030 |
0.1167981123 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007229_050 |
0.1296821143 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002046_060 |
0.1311959666 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 11 |
Hb_002391_320 |
0.1312250477 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 12 |
Hb_132840_160 |
0.1322278881 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000157_140 |
0.1331339978 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 14 |
Hb_005305_120 |
0.1333591515 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 15 |
Hb_000928_120 |
0.1335973273 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 16 |
Hb_000392_250 |
0.1347540675 |
- |
- |
PREDICTED: HVA22-like protein a [Jatropha curcas] |
| 17 |
Hb_002016_140 |
0.1357942634 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase BAH1-like 1 [Jatropha curcas] |
| 18 |
Hb_007537_030 |
0.1372591163 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_012762_040 |
0.1375114606 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 20 |
Hb_002486_080 |
0.1407555473 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |