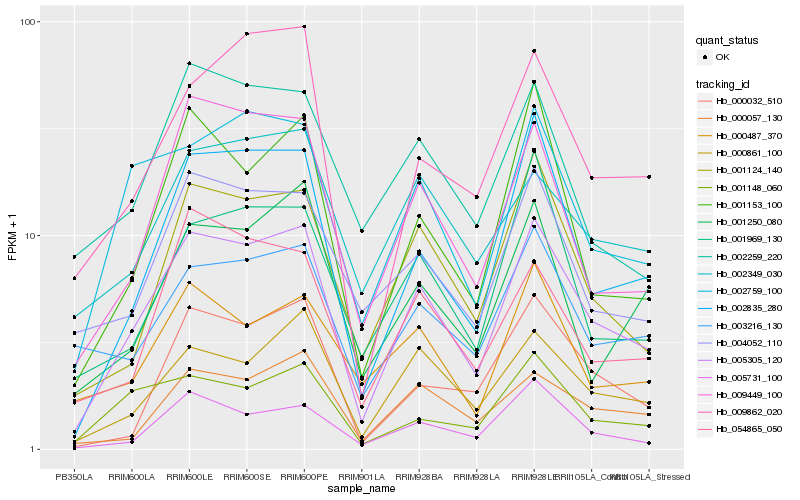
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005305_120 |
0.0 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 2 |
Hb_002835_280 |
0.1143667347 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.1 [Jatropha curcas] |
| 3 |
Hb_000861_100 |
0.1200681426 |
- |
- |
PREDICTED: probable fucosyltransferase 8 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001124_140 |
0.1206378549 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 5 |
Hb_002759_100 |
0.1237884687 |
- |
- |
hypothetical protein JCGZ_21777 [Jatropha curcas] |
| 6 |
Hb_009449_100 |
0.1245774224 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 1 [Jatropha curcas] |
| 7 |
Hb_005731_100 |
0.1331276125 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 8 |
Hb_000057_130 |
0.1333591515 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 9 |
Hb_001148_060 |
0.136346172 |
- |
- |
hypothetical protein JCGZ_00278 [Jatropha curcas] |
| 10 |
Hb_001153_100 |
0.1386911306 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 11 |
Hb_009862_020 |
0.1389667934 |
- |
- |
PREDICTED: uncharacterized protein LOC105650670 [Jatropha curcas] |
| 12 |
Hb_004052_110 |
0.1412809412 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000032_510 |
0.143352734 |
- |
- |
PREDICTED: SUN domain-containing protein 2 [Jatropha curcas] |
| 14 |
Hb_001250_080 |
0.1494657733 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g23390 [Jatropha curcas] |
| 15 |
Hb_003216_130 |
0.1497219675 |
- |
- |
PREDICTED: uncharacterized protein LOC105641500 [Jatropha curcas] |
| 16 |
Hb_001969_130 |
0.1497266102 |
- |
- |
PREDICTED: molybdate transporter 2 [Jatropha curcas] |
| 17 |
Hb_054865_050 |
0.1499976932 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 18 |
Hb_002349_030 |
0.1529142378 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002259_220 |
0.1531670004 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 20 |
Hb_000487_370 |
0.1536618581 |
- |
- |
- |