| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
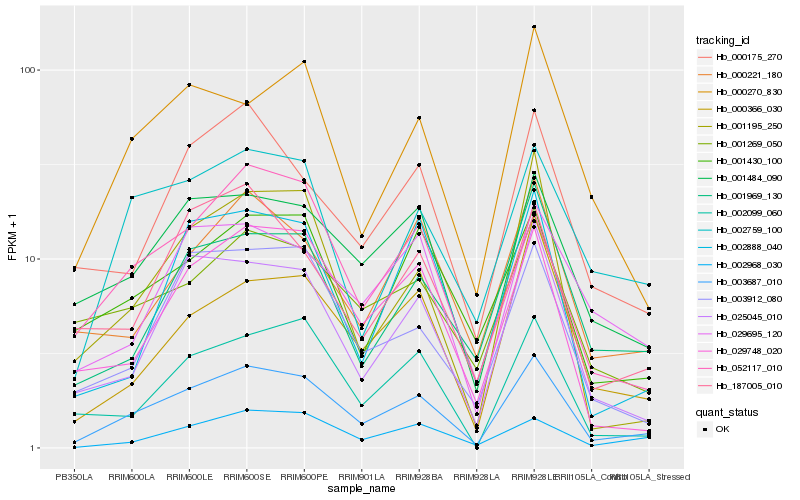
Hb_003687_010 |
0.0 |
- |
- |
Cysteine-rich RLK 29, putative [Theobroma cacao] |
| 2 |
Hb_025045_010 |
0.1319163632 |
- |
- |
catalytic, putative [Ricinus communis] |
| 3 |
Hb_001484_090 |
0.1341305674 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 4 |
Hb_029748_020 |
0.1386814293 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_002099_060 |
0.1404835178 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 6 |
Hb_000366_030 |
0.1442683262 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Jatropha curcas] |
| 7 |
Hb_002759_100 |
0.1446295322 |
- |
- |
hypothetical protein JCGZ_21777 [Jatropha curcas] |
| 8 |
Hb_003912_080 |
0.1447812938 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 9 |
Hb_002968_030 |
0.1456494108 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 10 |
Hb_001969_130 |
0.1475051357 |
- |
- |
PREDICTED: molybdate transporter 2 [Jatropha curcas] |
| 11 |
Hb_001195_250 |
0.1475259615 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 12 |
Hb_000175_270 |
0.1486665473 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 13 |
Hb_052117_010 |
0.1491111196 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_000221_180 |
0.1516650916 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 15 |
Hb_000270_830 |
0.1529999398 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_001430_100 |
0.1536856387 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_029695_120 |
0.155473441 |
- |
- |
hypothetical protein POPTR_0002s05320g [Populus trichocarpa] |
| 18 |
Hb_187005_010 |
0.1558592795 |
transcription factor |
TF Family: MYB-related |
PREDICTED: 1-phosphatidylinositol 3-phosphate 5-kinase [Jatropha curcas] |
| 19 |
Hb_001269_050 |
0.1565989639 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |
| 20 |
Hb_002888_040 |
0.1570881695 |
- |
- |
hypothetical protein B456_009G267200 [Gossypium raimondii] |