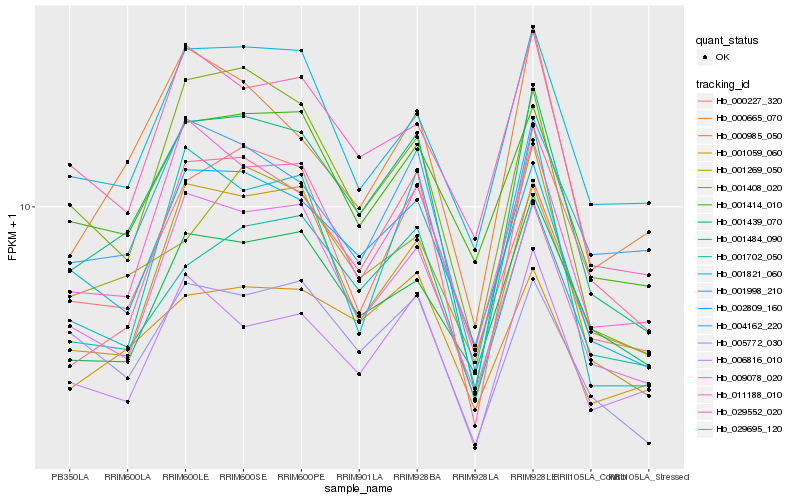
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001484_090 |
0.0 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_029695_120 |
0.0953258855 |
- |
- |
hypothetical protein POPTR_0002s05320g [Populus trichocarpa] |
| 3 |
Hb_002809_160 |
0.0971047454 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 4 |
Hb_001702_050 |
0.1000803228 |
- |
- |
PREDICTED: probable protein S-acyltransferase 19 isoform X1 [Jatropha curcas] |
| 5 |
Hb_009078_020 |
0.1023605256 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 6 |
Hb_001059_060 |
0.1069439277 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |
| 7 |
Hb_011188_010 |
0.1085675969 |
- |
- |
PREDICTED: uncharacterized protein LOC105642145 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001408_020 |
0.1110948516 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 9 |
Hb_000985_050 |
0.113363973 |
- |
- |
PREDICTED: uncharacterized protein LOC105647985 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005772_030 |
0.1138559044 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |
| 11 |
Hb_000665_070 |
0.1142281152 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_006816_010 |
0.114493468 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001998_210 |
0.1167796916 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_001269_050 |
0.1169759616 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |
| 15 |
Hb_001414_010 |
0.1186933402 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 16 |
Hb_004162_220 |
0.1204166633 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000227_320 |
0.1204719074 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_001439_070 |
0.1224034616 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 19 |
Hb_029552_020 |
0.1231801804 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001821_060 |
0.1264155994 |
- |
- |
PREDICTED: C-type lectin receptor-like tyrosine-protein kinase At1g52310 [Jatropha curcas] |