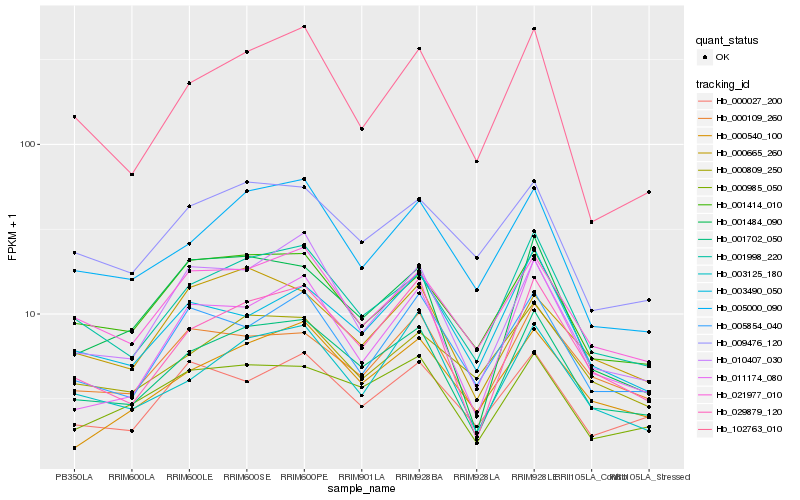
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001702_050 |
0.0 |
- |
- |
PREDICTED: probable protein S-acyltransferase 19 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001998_220 |
0.0709223838 |
- |
- |
PREDICTED: isoamylase 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_029879_120 |
0.0723517534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000985_050 |
0.0878934663 |
- |
- |
PREDICTED: uncharacterized protein LOC105647985 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005000_090 |
0.0962247164 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 6 |
Hb_021977_010 |
0.0964725169 |
- |
- |
PREDICTED: leukotriene A-4 hydrolase homolog [Jatropha curcas] |
| 7 |
Hb_003125_180 |
0.0992227468 |
- |
- |
hypothetical protein JCGZ_26440 [Jatropha curcas] |
| 8 |
Hb_001484_090 |
0.1000803228 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_010407_030 |
0.1001887303 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 10 |
Hb_000109_260 |
0.1003682703 |
- |
- |
PREDICTED: WAT1-related protein At3g28050-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_001414_010 |
0.101553013 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 12 |
Hb_011174_080 |
0.1035570833 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 13 |
Hb_000540_100 |
0.1039611222 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g60050 [Jatropha curcas] |
| 14 |
Hb_102763_010 |
0.1055086882 |
- |
- |
PREDICTED: heat shock cognate protein 80 [Jatropha curcas] |
| 15 |
Hb_009476_120 |
0.1082067013 |
- |
- |
Clathrin heavy chain 1 [Glycine soja] |
| 16 |
Hb_000809_250 |
0.1110297013 |
- |
- |
PREDICTED: uncharacterized protein LOC105639683 [Jatropha curcas] |
| 17 |
Hb_005854_040 |
0.1112246821 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000027_200 |
0.111265223 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 19 |
Hb_000665_260 |
0.1115170969 |
- |
- |
PREDICTED: uncharacterized protein LOC105637620 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003490_050 |
0.1117390671 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic [Jatropha curcas] |