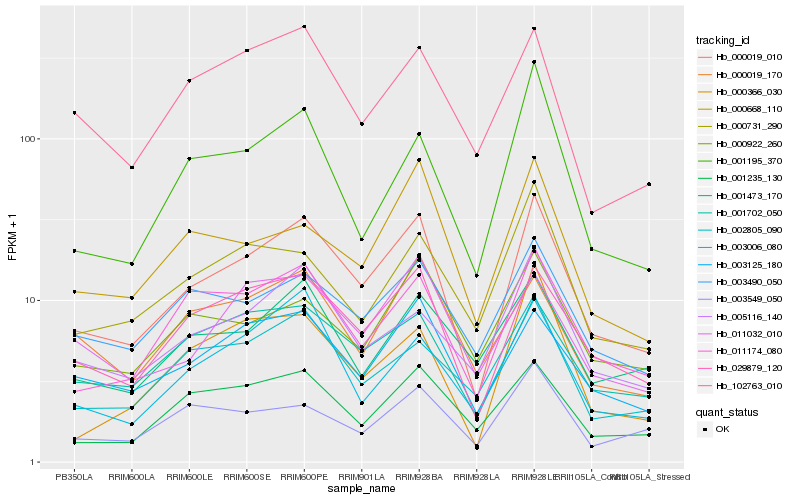
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000019_010 |
0.0 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Jatropha curcas] |
| 2 |
Hb_029879_120 |
0.0903765497 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_011174_080 |
0.1119416518 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 4 |
Hb_003125_180 |
0.1236171011 |
- |
- |
hypothetical protein JCGZ_26440 [Jatropha curcas] |
| 5 |
Hb_001702_050 |
0.1263919169 |
- |
- |
PREDICTED: probable protein S-acyltransferase 19 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001235_130 |
0.1324274159 |
- |
- |
- |
| 7 |
Hb_003490_050 |
0.1353140571 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic [Jatropha curcas] |
| 8 |
Hb_005116_140 |
0.1363219851 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 9 |
Hb_002805_090 |
0.1415837157 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 10 |
Hb_000668_110 |
0.1416930272 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 11 |
Hb_011032_010 |
0.1418881936 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 12 |
Hb_003006_080 |
0.1421555406 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_001195_370 |
0.1431015031 |
- |
- |
- |
| 14 |
Hb_102763_010 |
0.1450926226 |
- |
- |
PREDICTED: heat shock cognate protein 80 [Jatropha curcas] |
| 15 |
Hb_000922_260 |
0.1455359274 |
- |
- |
PREDICTED: pantothenate kinase 2 [Jatropha curcas] |
| 16 |
Hb_001473_170 |
0.1459583807 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 17 |
Hb_000366_030 |
0.1473156247 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Jatropha curcas] |
| 18 |
Hb_000731_290 |
0.1490132223 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_000019_170 |
0.1493804142 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 20 |
Hb_003549_050 |
0.1497467598 |
- |
- |
PREDICTED: zinc phosphodiesterase ELAC protein 2-like [Jatropha curcas] |