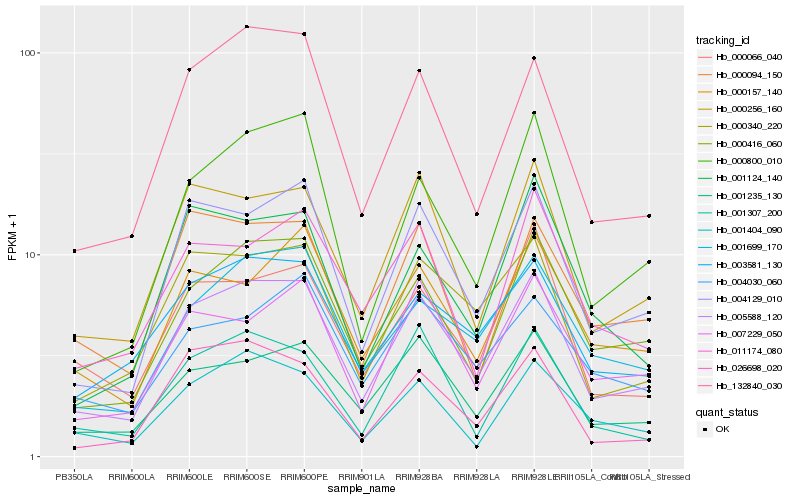
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005588_120 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004129_010 |
0.0749372905 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 3 |
Hb_000416_060 |
0.0833174017 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 4 |
Hb_132840_030 |
0.085959422 |
transcription factor |
TF Family: MYB-related |
lhy1 [Populus tremula] |
| 5 |
Hb_000256_160 |
0.0884051492 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3-like isoform X1 [Glycine max] |
| 6 |
Hb_001235_130 |
0.0905781421 |
- |
- |
- |
| 7 |
Hb_000340_220 |
0.0957541263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001699_170 |
0.0986678529 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003581_130 |
0.1000002535 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_000800_010 |
0.1020193105 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 11 |
Hb_007229_050 |
0.1081770033 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000066_040 |
0.1105973218 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 13 |
Hb_000157_140 |
0.1133571695 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 14 |
Hb_026698_020 |
0.1139594243 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 15 |
Hb_001404_090 |
0.1141879695 |
- |
- |
PREDICTED: kinesin-like protein NACK2 [Camelina sativa] |
| 16 |
Hb_000094_150 |
0.1142526166 |
- |
- |
- |
| 17 |
Hb_001124_140 |
0.11824149 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 18 |
Hb_001307_200 |
0.1193021833 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |
| 19 |
Hb_011174_080 |
0.1215096615 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 20 |
Hb_004030_060 |
0.121689038 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |