| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
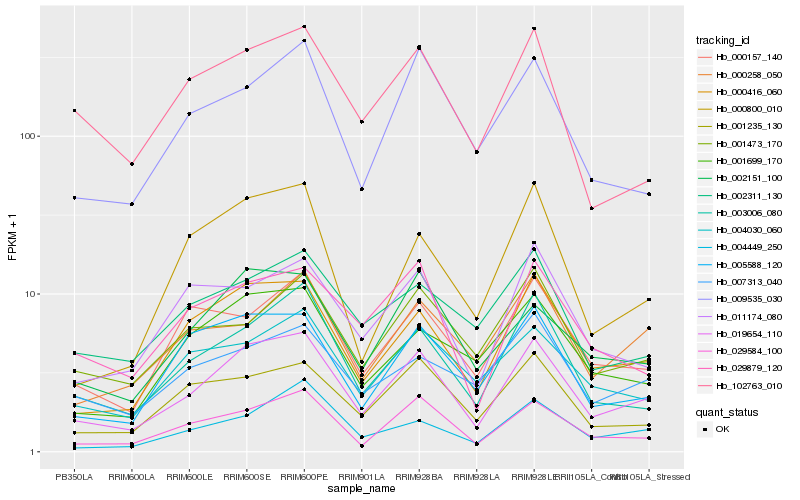
Hb_019654_110 |
0.0 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Jatropha curcas] |
| 2 |
Hb_000416_060 |
0.0984226173 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 3 |
Hb_029584_100 |
0.105702068 |
- |
- |
Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase [Gossypium arboreum] |
| 4 |
Hb_003006_080 |
0.1130072458 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_002151_100 |
0.125184086 |
- |
- |
PREDICTED: uncharacterized protein LOC105637951 [Jatropha curcas] |
| 6 |
Hb_005588_120 |
0.1284225939 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001235_130 |
0.1290620631 |
- |
- |
- |
| 8 |
Hb_009535_030 |
0.1318323284 |
- |
- |
CP2 [Hevea brasiliensis] |
| 9 |
Hb_001699_170 |
0.135093481 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_029879_120 |
0.1372912433 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004030_060 |
0.1385311177 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 12 |
Hb_000800_010 |
0.1390465508 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 13 |
Hb_000157_140 |
0.1402030611 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 14 |
Hb_001473_170 |
0.1406047147 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 15 |
Hb_004449_250 |
0.1408566987 |
- |
- |
PREDICTED: protein EI24 homolog [Jatropha curcas] |
| 16 |
Hb_102763_010 |
0.141846367 |
- |
- |
PREDICTED: heat shock cognate protein 80 [Jatropha curcas] |
| 17 |
Hb_007313_040 |
0.1419061165 |
- |
- |
hypothetical protein JCGZ_23401 [Jatropha curcas] |
| 18 |
Hb_000258_050 |
0.1419775632 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002311_130 |
0.1428449761 |
- |
- |
PREDICTED: importin subunit alpha-2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_011174_080 |
0.1429610531 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |