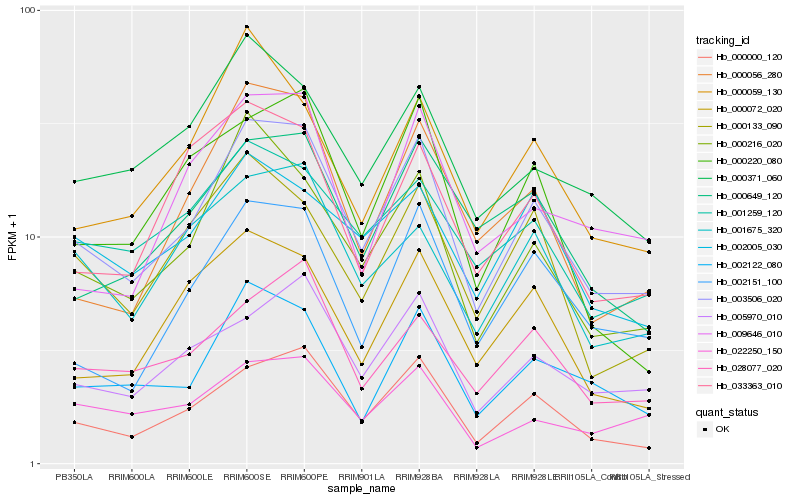
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003506_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105125881 isoform X1 [Populus euphratica] |
| 2 |
Hb_000000_120 |
0.0720286851 |
- |
- |
hypothetical protein EUGRSUZ_A004731, partial [Eucalyptus grandis] |
| 3 |
Hb_000216_020 |
0.1018418165 |
- |
- |
PREDICTED: benzaldehyde dehydrogenase (NAD(+))-like [Jatropha curcas] |
| 4 |
Hb_005970_010 |
0.1029876195 |
- |
- |
PREDICTED: probable isoprenylcysteine alpha-carbonyl methylesterase ICMEL1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002122_080 |
0.1047032984 |
- |
- |
- |
| 6 |
Hb_002151_100 |
0.1085365978 |
- |
- |
PREDICTED: uncharacterized protein LOC105637951 [Jatropha curcas] |
| 7 |
Hb_028077_020 |
0.1144637426 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001675_320 |
0.1156452231 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 9 |
Hb_000056_280 |
0.1175146148 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 10 |
Hb_000371_060 |
0.1192733133 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 11 |
Hb_022250_150 |
0.1196230611 |
- |
- |
PREDICTED: uncharacterized protein LOC105648687 [Jatropha curcas] |
| 12 |
Hb_000649_120 |
0.1197406725 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_001259_120 |
0.1199306666 |
- |
- |
hypothetical protein EUGRSUZ_C043222, partial [Eucalyptus grandis] |
| 14 |
Hb_000072_020 |
0.1218569873 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000133_090 |
0.1234011245 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 16 |
Hb_009646_010 |
0.1268750458 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 17 |
Hb_000220_080 |
0.1269879032 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_000059_130 |
0.1273418627 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 19 |
Hb_002005_030 |
0.1274112779 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 20 |
Hb_033363_010 |
0.1276030299 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |