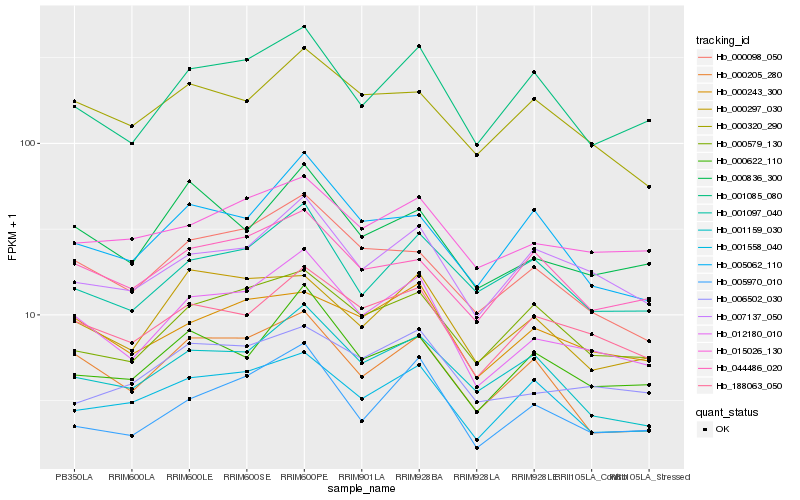
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012180_010 |
0.0 |
- |
- |
PREDICTED: (S)-ureidoglycine aminohydrolase [Jatropha curcas] |
| 2 |
Hb_188063_050 |
0.0917706101 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 3 |
Hb_000098_050 |
0.0942076816 |
- |
- |
BnaCnng11900D [Brassica napus] |
| 4 |
Hb_001085_080 |
0.0964052994 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 5 |
Hb_007137_050 |
0.0988745864 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 6 |
Hb_005970_010 |
0.0990192228 |
- |
- |
PREDICTED: probable isoprenylcysteine alpha-carbonyl methylesterase ICMEL1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000622_110 |
0.1024810336 |
- |
- |
cmp-sialic acid transporter, putative [Ricinus communis] |
| 8 |
Hb_000836_300 |
0.1031929523 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |
| 9 |
Hb_001159_030 |
0.1062206849 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 10 |
Hb_000243_300 |
0.1062326491 |
- |
- |
catalytic, putative [Ricinus communis] |
| 11 |
Hb_000205_280 |
0.1089508123 |
- |
- |
PREDICTED: inositol phosphorylceramide glucuronosyltransferase 1 [Jatropha curcas] |
| 12 |
Hb_015026_130 |
0.1106553321 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 13 |
Hb_001097_040 |
0.1108131499 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 14 |
Hb_000579_130 |
0.1111848541 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_001558_040 |
0.1123620965 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 2 [Jatropha curcas] |
| 16 |
Hb_005062_110 |
0.1139151139 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 1 [Jatropha curcas] |
| 17 |
Hb_000297_030 |
0.1142323334 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000320_290 |
0.1146058159 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |
| 19 |
Hb_006502_030 |
0.115679046 |
- |
- |
PREDICTED: probable kinetochore protein ndc80 [Jatropha curcas] |
| 20 |
Hb_044486_020 |
0.1166442607 |
- |
- |
CASTOR protein [Glycine max] |