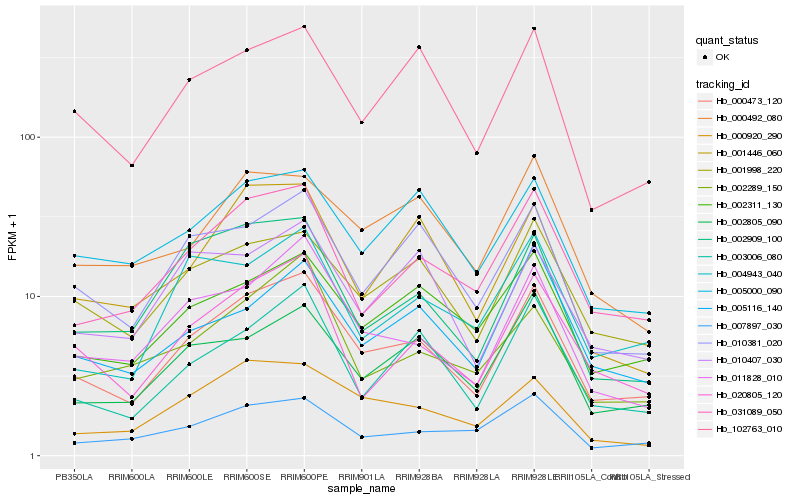
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000473_120 |
0.0 |
- |
- |
hypothetical protein JCGZ_16177 [Jatropha curcas] |
| 2 |
Hb_031089_050 |
0.0981057753 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_011828_010 |
0.1031980324 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 4 |
Hb_001998_220 |
0.1087344146 |
- |
- |
PREDICTED: isoamylase 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_020805_120 |
0.1122347436 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 6 |
Hb_003006_080 |
0.1124241782 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_002311_130 |
0.1144010233 |
- |
- |
PREDICTED: importin subunit alpha-2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_010381_020 |
0.1160782711 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 9 |
Hb_000920_290 |
0.116218879 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 10 |
Hb_102763_010 |
0.1187125891 |
- |
- |
PREDICTED: heat shock cognate protein 80 [Jatropha curcas] |
| 11 |
Hb_002805_090 |
0.1195673419 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 12 |
Hb_007897_030 |
0.1219296439 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR2 [Jatropha curcas] |
| 13 |
Hb_002909_100 |
0.1225240401 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 14 |
Hb_005000_090 |
0.1271690835 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 15 |
Hb_004943_040 |
0.1293195233 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g30440 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001446_060 |
0.1294431465 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_005116_140 |
0.1314780916 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 18 |
Hb_000492_080 |
0.1315611068 |
- |
- |
PREDICTED: probable sucrose-phosphate synthase 1 [Jatropha curcas] |
| 19 |
Hb_002289_150 |
0.1338944699 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 3 [UDP-forming]-like [Jatropha curcas] |
| 20 |
Hb_010407_030 |
0.1342954304 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |