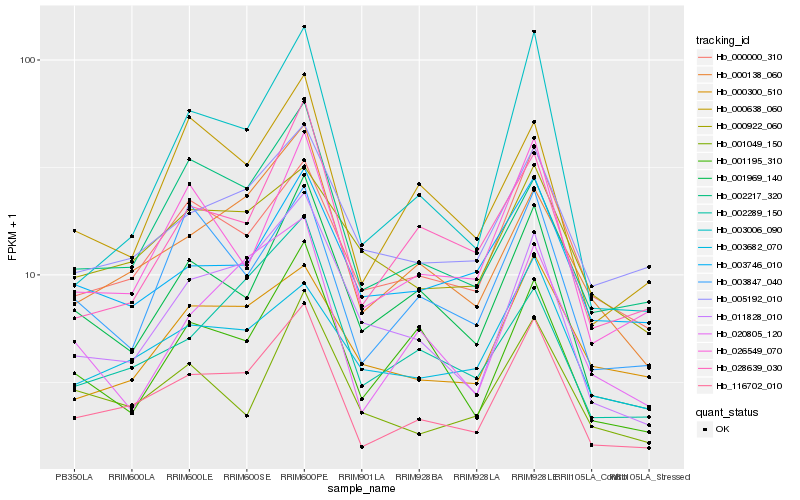
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_116702_010 |
0.0 |
- |
- |
Leucine-rich repeat protein kinase family protein [Theobroma cacao] |
| 2 |
Hb_000000_310 |
0.1027703002 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 3 |
Hb_000138_060 |
0.1093029069 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 4 |
Hb_026549_070 |
0.1137854294 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 5 |
Hb_005192_010 |
0.1149482876 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 6 |
Hb_001969_140 |
0.116130252 |
- |
- |
PREDICTED: uncharacterized protein LOC105634929 [Jatropha curcas] |
| 7 |
Hb_011828_010 |
0.1215315421 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 8 |
Hb_003682_070 |
0.125270126 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003746_010 |
0.1287476754 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_028639_030 |
0.1288492741 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 2-like [Jatropha curcas] |
| 11 |
Hb_002217_320 |
0.1290478154 |
- |
- |
PREDICTED: probable O-acetyltransferase CAS1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_003847_040 |
0.1305882725 |
- |
- |
adenosine diphosphatase, putative [Ricinus communis] |
| 13 |
Hb_001049_150 |
0.1308256547 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 14 |
Hb_001195_310 |
0.1314430466 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_000638_060 |
0.131720555 |
- |
- |
PREDICTED: probable methyltransferase PMT26 [Jatropha curcas] |
| 16 |
Hb_002289_150 |
0.1347428289 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 3 [UDP-forming]-like [Jatropha curcas] |
| 17 |
Hb_000922_060 |
0.1347711072 |
- |
- |
kinesin light chain, putative [Ricinus communis] |
| 18 |
Hb_003006_090 |
0.1349524273 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 19 |
Hb_020805_120 |
0.1354958864 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 20 |
Hb_000300_510 |
0.1361220813 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |