| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
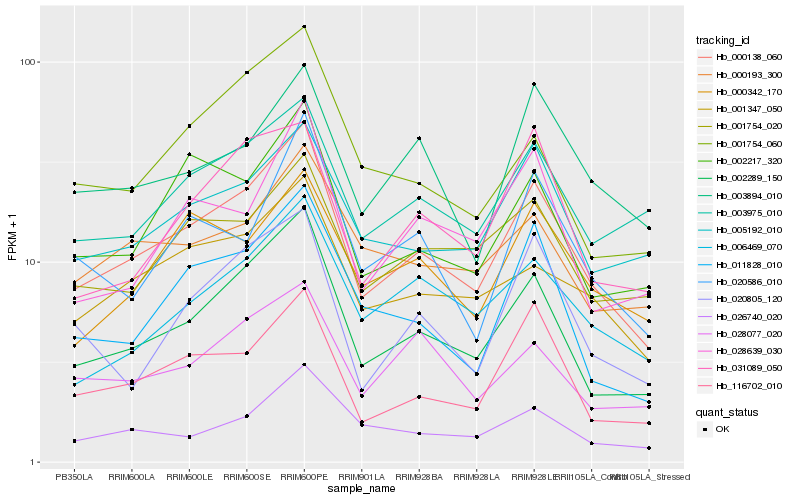
Hb_000138_060 |
0.0 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 2 |
Hb_002289_150 |
0.0669641431 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 3 [UDP-forming]-like [Jatropha curcas] |
| 3 |
Hb_116702_010 |
0.1093029069 |
- |
- |
Leucine-rich repeat protein kinase family protein [Theobroma cacao] |
| 4 |
Hb_011828_010 |
0.1117677689 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 5 |
Hb_001347_050 |
0.1132843886 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 6 |
Hb_002217_320 |
0.115184447 |
- |
- |
PREDICTED: probable O-acetyltransferase CAS1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_005192_010 |
0.1212492428 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 8 |
Hb_003975_010 |
0.1228456727 |
- |
- |
PREDICTED: protein RTF2 homolog [Jatropha curcas] |
| 9 |
Hb_026740_020 |
0.1229873124 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 10 |
Hb_006469_070 |
0.1245140502 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g22040 [Jatropha curcas] |
| 11 |
Hb_020805_120 |
0.1248601402 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 12 |
Hb_001754_020 |
0.1264214173 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 13 |
Hb_028639_030 |
0.1276843107 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 2-like [Jatropha curcas] |
| 14 |
Hb_031089_050 |
0.128558424 |
- |
- |
kinase, putative [Ricinus communis] |
| 15 |
Hb_001754_060 |
0.1289302889 |
- |
- |
Xyloglucan 6-xylosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000193_300 |
0.1300008419 |
- |
- |
PREDICTED: putative glycosyltransferase 5 [Jatropha curcas] |
| 17 |
Hb_020586_010 |
0.1319933929 |
- |
- |
Alpha-N-arabinofuranosidase 1 precursor, putative [Ricinus communis] |
| 18 |
Hb_000342_170 |
0.1322279222 |
- |
- |
PREDICTED: uncharacterized protein LOC100807540 isoform X2 [Glycine max] |
| 19 |
Hb_028077_020 |
0.1326571134 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003894_010 |
0.135081729 |
- |
- |
unnamed protein product [Vitis vinifera] |