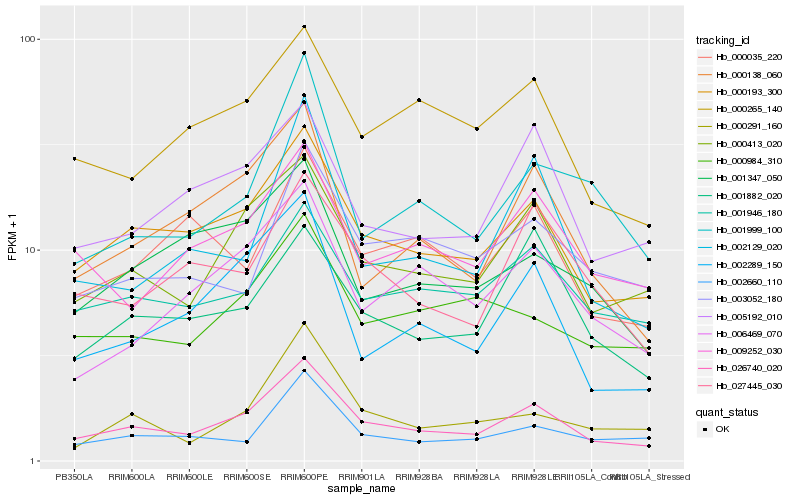
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026740_020 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 2 |
Hb_000193_300 |
0.0745132877 |
- |
- |
PREDICTED: putative glycosyltransferase 5 [Jatropha curcas] |
| 3 |
Hb_000413_020 |
0.0901444931 |
- |
- |
PREDICTED: U-box domain-containing protein 8 [Jatropha curcas] |
| 4 |
Hb_000138_060 |
0.1229873124 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 5 |
Hb_002289_150 |
0.1245069172 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 3 [UDP-forming]-like [Jatropha curcas] |
| 6 |
Hb_005192_010 |
0.1268567056 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 7 |
Hb_000265_140 |
0.1272679766 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 12 [Jatropha curcas] |
| 8 |
Hb_001946_180 |
0.1278451682 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_027445_030 |
0.1303292725 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |
| 10 |
Hb_009252_030 |
0.1310613412 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 11 |
Hb_001882_020 |
0.1328043117 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 12 |
Hb_006469_070 |
0.1329430397 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g22040 [Jatropha curcas] |
| 13 |
Hb_002660_110 |
0.1348053805 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002129_020 |
0.1352589033 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 15 |
Hb_000984_310 |
0.1373276868 |
- |
- |
Hydrolase, hydrolyzing O-glycosyl compounds, putative [Theobroma cacao] |
| 16 |
Hb_001999_100 |
0.1399763432 |
- |
- |
PREDICTED: UDP-glycosyltransferase 71D1-like [Jatropha curcas] |
| 17 |
Hb_000035_220 |
0.1404561505 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 8 [Jatropha curcas] |
| 18 |
Hb_001347_050 |
0.1406572462 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 19 |
Hb_003052_180 |
0.1410745575 |
- |
- |
Diacylglycerol Cholinephosphotransferase [Ricinus communis] |
| 20 |
Hb_000291_160 |
0.144408207 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |