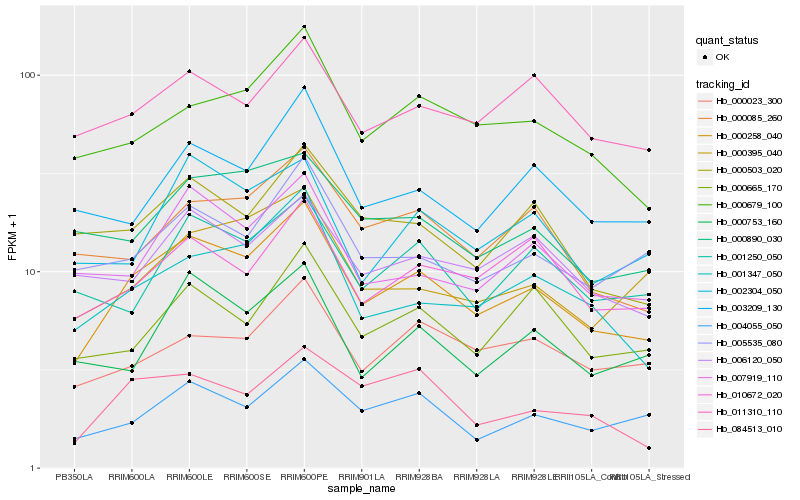
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000258_040 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 2 |
Hb_001347_050 |
0.1076478078 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 3 |
Hb_004055_050 |
0.112522262 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 4 |
Hb_000679_100 |
0.1126468678 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 5 |
Hb_007919_110 |
0.1163745415 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 6 |
Hb_000085_260 |
0.1175280543 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 7 |
Hb_010672_020 |
0.1181779805 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_005535_080 |
0.1207826702 |
- |
- |
hypothetical protein CICLE_v10012766mg [Citrus clementina] |
| 9 |
Hb_003209_130 |
0.1209738357 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 10 |
Hb_000395_040 |
0.1220995254 |
- |
- |
PREDICTED: uncharacterized protein LOC105645576 [Jatropha curcas] |
| 11 |
Hb_000503_020 |
0.1226526111 |
- |
- |
PREDICTED: calcium-dependent protein kinase 13 [Jatropha curcas] |
| 12 |
Hb_000665_170 |
0.1226718477 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 13 |
Hb_006120_050 |
0.1236856272 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 14 |
Hb_011310_110 |
0.1238117105 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 15 |
Hb_084513_010 |
0.1243157099 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 5 [Jatropha curcas] |
| 16 |
Hb_000023_300 |
0.1246730467 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 17 |
Hb_000753_160 |
0.1247257922 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 18 |
Hb_002304_050 |
0.1247941754 |
- |
- |
pyruvate kinase, putative [Ricinus communis] |
| 19 |
Hb_000890_030 |
0.1248299835 |
- |
- |
plastid CUL1 [Hevea brasiliensis] |
| 20 |
Hb_001250_050 |
0.1250768016 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |