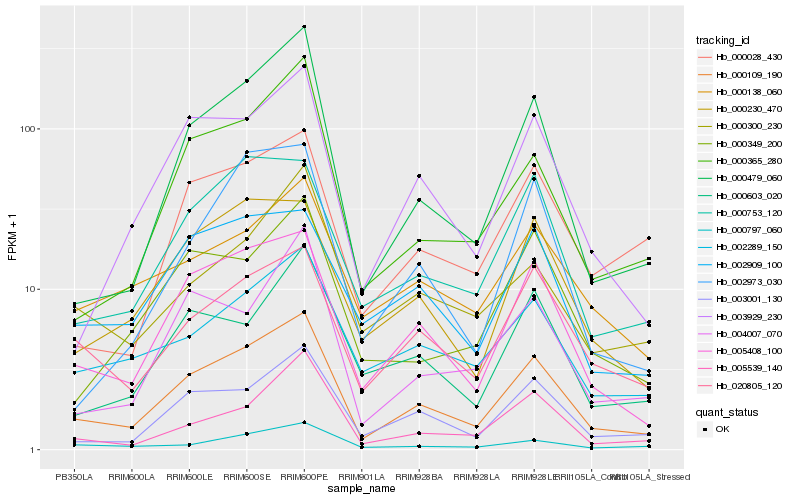
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000109_190 |
0.0 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 2 |
Hb_005408_100 |
0.0932427676 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 3 |
Hb_003001_130 |
0.1146360388 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000479_060 |
0.1167228905 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1 [Vitis vinifera] |
| 5 |
Hb_020805_120 |
0.1262840282 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 6 |
Hb_003929_230 |
0.1278715491 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 5 [Nelumbo nucifera] |
| 7 |
Hb_000365_280 |
0.1330814317 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 8 |
Hb_000753_120 |
0.1352486313 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 9 |
Hb_002289_150 |
0.1386068283 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 3 [UDP-forming]-like [Jatropha curcas] |
| 10 |
Hb_000230_470 |
0.1407156623 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 11 |
Hb_005539_140 |
0.145086666 |
- |
- |
PREDICTED: U-box domain-containing protein 19-like [Jatropha curcas] |
| 12 |
Hb_004007_070 |
0.1492072424 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 13 |
Hb_000797_060 |
0.1508758308 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 14 |
Hb_002909_100 |
0.1516518099 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 15 |
Hb_000603_020 |
0.1519404023 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000138_060 |
0.1531560273 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 17 |
Hb_002973_030 |
0.153952521 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 18 |
Hb_000349_200 |
0.1542462976 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g12500 [Jatropha curcas] |
| 19 |
Hb_000028_430 |
0.1544338572 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000300_230 |
0.1558952085 |
- |
- |
PREDICTED: cation/calcium exchanger 2 [Jatropha curcas] |