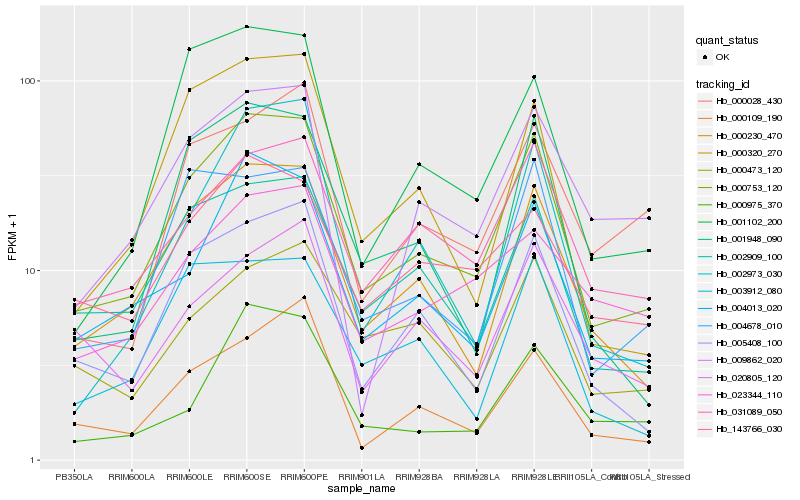
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000753_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 2 |
Hb_000230_470 |
0.0908890586 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 3 |
Hb_004013_020 |
0.107690612 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |
| 4 |
Hb_002909_100 |
0.1080138512 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 5 |
Hb_005408_100 |
0.1115487411 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 6 |
Hb_031089_050 |
0.1174088408 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_001948_090 |
0.1278837508 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0005s14540g [Populus trichocarpa] |
| 8 |
Hb_004678_010 |
0.1290271266 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 9 |
Hb_143766_030 |
0.1330224962 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_000320_270 |
0.1337033113 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001102_200 |
0.133725147 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 12 |
Hb_000109_190 |
0.1352486313 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 13 |
Hb_003912_080 |
0.1400907634 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 14 |
Hb_002973_030 |
0.1435835265 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 15 |
Hb_009862_020 |
0.1445928237 |
- |
- |
PREDICTED: uncharacterized protein LOC105650670 [Jatropha curcas] |
| 16 |
Hb_000473_120 |
0.1449784119 |
- |
- |
hypothetical protein JCGZ_16177 [Jatropha curcas] |
| 17 |
Hb_000975_370 |
0.1460187036 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB59 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000028_430 |
0.1480246627 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 19 |
Hb_020805_120 |
0.1514618898 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 20 |
Hb_023344_110 |
0.1523480775 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 [Jatropha curcas] |