| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
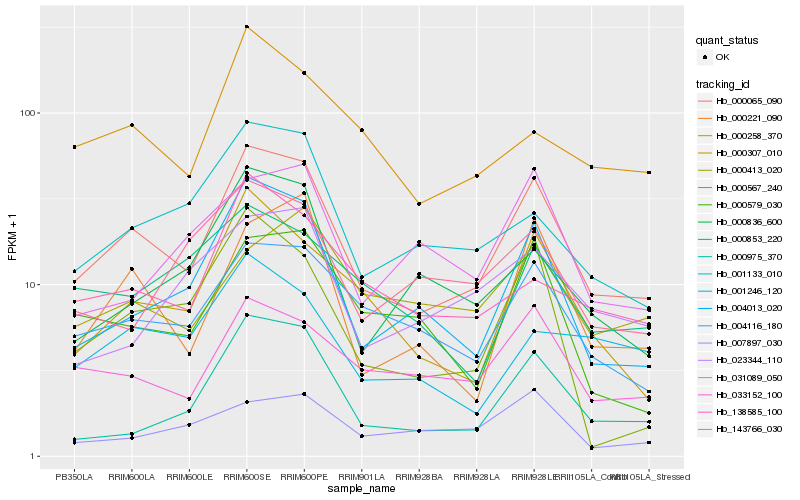
Hb_000065_090 |
0.0 |
- |
- |
potassium channel tetramerization domain-containing protein, putative [Ricinus communis] |
| 2 |
Hb_004013_020 |
0.1321607204 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |
| 3 |
Hb_004116_180 |
0.1362333738 |
- |
- |
PREDICTED: uncharacterized protein LOC105646147 [Jatropha curcas] |
| 4 |
Hb_000307_010 |
0.1462937191 |
- |
- |
PREDICTED: ganglioside-induced differentiation-associated protein 2-like [Jatropha curcas] |
| 5 |
Hb_033152_100 |
0.1467004819 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000221_090 |
0.1490242152 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-6-like [Jatropha curcas] |
| 7 |
Hb_001133_010 |
0.1589738284 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000975_370 |
0.1595162595 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB59 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000258_370 |
0.165746645 |
- |
- |
hypothetical protein [Ricinus communis] |
| 10 |
Hb_138585_100 |
0.1665393612 |
- |
- |
PREDICTED: uncharacterized protein LOC105648740 [Jatropha curcas] |
| 11 |
Hb_000579_030 |
0.1704119498 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 3 [Jatropha curcas] |
| 12 |
Hb_000836_600 |
0.1728375317 |
- |
- |
PREDICTED: delta(8)-fatty-acid desaturase 2 [Jatropha curcas] |
| 13 |
Hb_000567_240 |
0.1754132349 |
- |
- |
PREDICTED: F-box/LRR-repeat MAX2 homolog A-like [Jatropha curcas] |
| 14 |
Hb_000413_020 |
0.1812653061 |
- |
- |
PREDICTED: U-box domain-containing protein 8 [Jatropha curcas] |
| 15 |
Hb_001246_120 |
0.1813120746 |
- |
- |
PREDICTED: uncharacterized protein LOC105643270 [Jatropha curcas] |
| 16 |
Hb_000853_220 |
0.1813510299 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 17 |
Hb_031089_050 |
0.1816073572 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_023344_110 |
0.1829250108 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 [Jatropha curcas] |
| 19 |
Hb_007897_030 |
0.1835022775 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR2 [Jatropha curcas] |
| 20 |
Hb_143766_030 |
0.1840979251 |
- |
- |
protein with unknown function [Ricinus communis] |