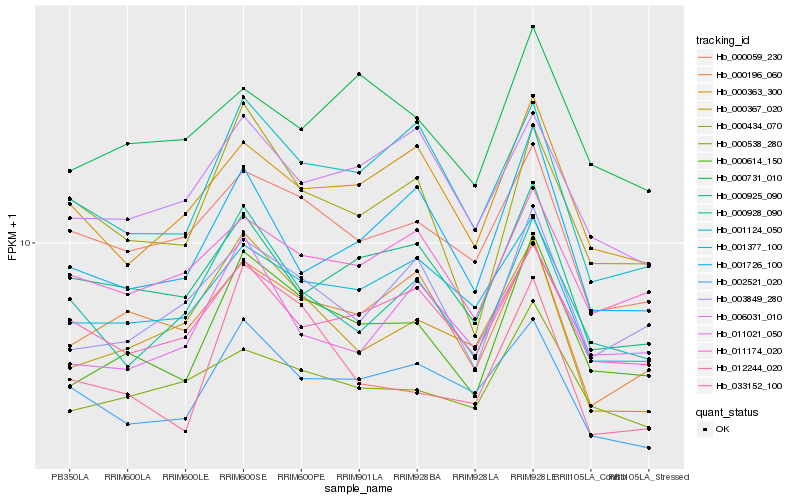
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000614_150 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit B-like [Jatropha curcas] |
| 2 |
Hb_003849_280 |
0.095938864 |
- |
- |
PREDICTED: uncharacterized protein LOC105644977 [Jatropha curcas] |
| 3 |
Hb_000434_070 |
0.1049815193 |
- |
- |
PREDICTED: uncharacterized transmembrane protein DDB_G0289901 [Jatropha curcas] |
| 4 |
Hb_000538_280 |
0.1083208585 |
- |
- |
PREDICTED: uncharacterized protein LOC105642112 [Jatropha curcas] |
| 5 |
Hb_000196_060 |
0.1107830003 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 6 |
Hb_001124_050 |
0.1110935231 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 7 |
Hb_001377_100 |
0.1120804586 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000059_230 |
0.1140865393 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 9 |
Hb_011021_050 |
0.1144255925 |
- |
- |
PREDICTED: uncharacterized protein LOC105644387 isoform X1 [Jatropha curcas] |
| 10 |
Hb_012244_020 |
0.1147002201 |
- |
- |
calpain, putative [Ricinus communis] |
| 11 |
Hb_000367_020 |
0.1147978714 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 12 |
Hb_000928_090 |
0.1152615042 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 13 |
Hb_011174_020 |
0.1161096436 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 14 |
Hb_000363_300 |
0.1161514377 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001726_100 |
0.1161775863 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 16 |
Hb_033152_100 |
0.1164662985 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000731_010 |
0.1166471542 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_006031_010 |
0.1169106122 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X3 [Jatropha curcas] |
| 19 |
Hb_000925_090 |
0.1173063673 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002521_020 |
0.1179853338 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03100, mitochondrial-like isoform X2 [Jatropha curcas] |