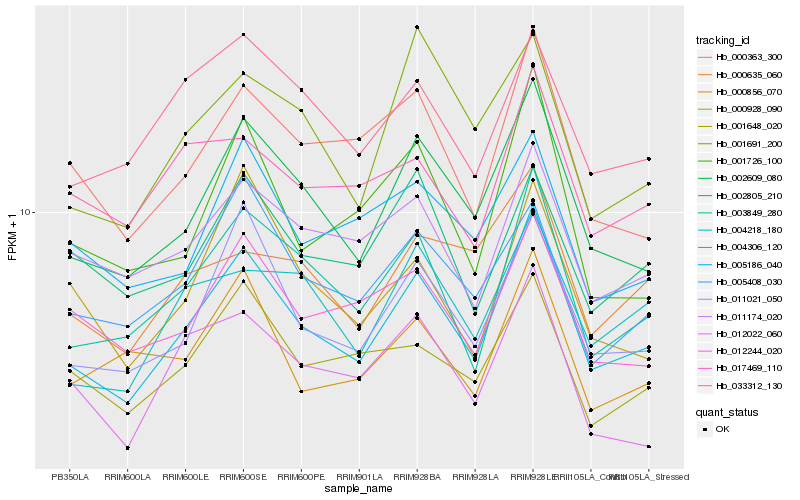
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004306_120 |
0.0 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_002609_080 |
0.0530583403 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_011021_050 |
0.0629295699 |
- |
- |
PREDICTED: uncharacterized protein LOC105644387 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003849_280 |
0.0631165678 |
- |
- |
PREDICTED: uncharacterized protein LOC105644977 [Jatropha curcas] |
| 5 |
Hb_000363_300 |
0.0791196835 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005408_030 |
0.0816504613 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 7 |
Hb_001648_020 |
0.0818853154 |
- |
- |
hypothetical protein POPTR_0007s07750g [Populus trichocarpa] |
| 8 |
Hb_011174_020 |
0.0820977354 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 9 |
Hb_004218_180 |
0.0837765364 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 10 |
Hb_012244_020 |
0.0839357114 |
- |
- |
calpain, putative [Ricinus communis] |
| 11 |
Hb_033312_130 |
0.0873081752 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 12 |
Hb_012022_060 |
0.0875288299 |
- |
- |
PREDICTED: uncharacterized protein LOC105646588 isoform X2 [Jatropha curcas] |
| 13 |
Hb_005186_040 |
0.0877127497 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002805_210 |
0.0878062544 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 15 |
Hb_001726_100 |
0.0888151042 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000856_070 |
0.0895840556 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 17 |
Hb_000928_090 |
0.0914335652 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 18 |
Hb_000635_060 |
0.0923343841 |
- |
- |
PREDICTED: uncharacterized protein LOC105644797 [Jatropha curcas] |
| 19 |
Hb_001691_200 |
0.0923991602 |
- |
- |
DNA-directed RNA polymerase II subunit RPB2 [Auxenochlorella protothecoides] |
| 20 |
Hb_017469_110 |
0.0926625719 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |