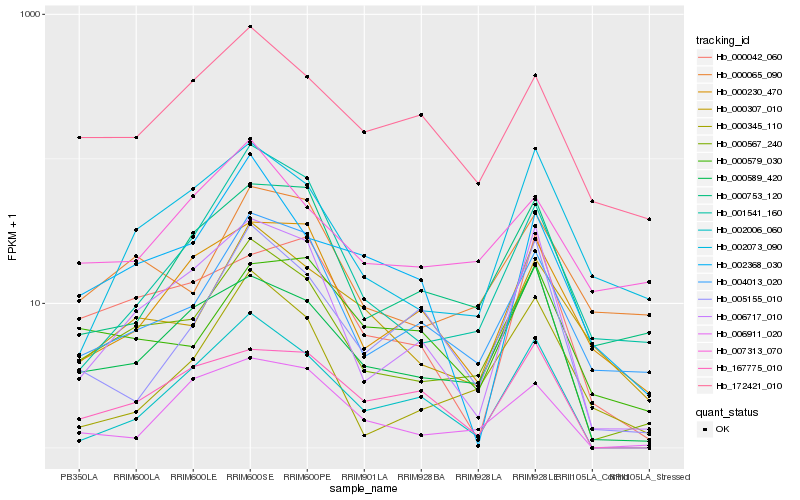
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000567_240 |
0.0 |
- |
- |
PREDICTED: F-box/LRR-repeat MAX2 homolog A-like [Jatropha curcas] |
| 2 |
Hb_000589_420 |
0.1373550524 |
- |
- |
PREDICTED: uncharacterized protein LOC105647273 [Jatropha curcas] |
| 3 |
Hb_006717_010 |
0.1392374359 |
- |
- |
PREDICTED: uncharacterized protein LOC105649338 [Jatropha curcas] |
| 4 |
Hb_004013_020 |
0.148305397 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |
| 5 |
Hb_000307_010 |
0.1701323391 |
- |
- |
PREDICTED: ganglioside-induced differentiation-associated protein 2-like [Jatropha curcas] |
| 6 |
Hb_000065_090 |
0.1754132349 |
- |
- |
potassium channel tetramerization domain-containing protein, putative [Ricinus communis] |
| 7 |
Hb_000753_120 |
0.1788761138 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 8 |
Hb_002073_090 |
0.1795877666 |
- |
- |
PREDICTED: copper transporter 6-like [Jatropha curcas] |
| 9 |
Hb_001541_160 |
0.1796994261 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 10 |
Hb_172421_010 |
0.1835465873 |
- |
- |
cysteine protease [Hevea brasiliensis] |
| 11 |
Hb_002006_060 |
0.1837146077 |
- |
- |
PREDICTED: oleosin 1-like [Jatropha curcas] |
| 12 |
Hb_000345_110 |
0.1904431345 |
- |
- |
PREDICTED: uncharacterized protein LOC105647507 [Jatropha curcas] |
| 13 |
Hb_000230_470 |
0.1935985172 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 14 |
Hb_005155_010 |
0.1963008366 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 15 |
Hb_006911_020 |
0.1975527091 |
- |
- |
CC-NBS-LRR protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_167775_010 |
0.1996977008 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 17 |
Hb_007313_070 |
0.2004916986 |
- |
- |
PREDICTED: gibberellin receptor GID1C [Jatropha curcas] |
| 18 |
Hb_002368_030 |
0.2017028761 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein GAIP-B [Jatropha curcas] |
| 19 |
Hb_000579_030 |
0.201980166 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 3 [Jatropha curcas] |
| 20 |
Hb_000042_060 |
0.2038415767 |
- |
- |
PREDICTED: uncharacterized protein LOC105632794 [Jatropha curcas] |