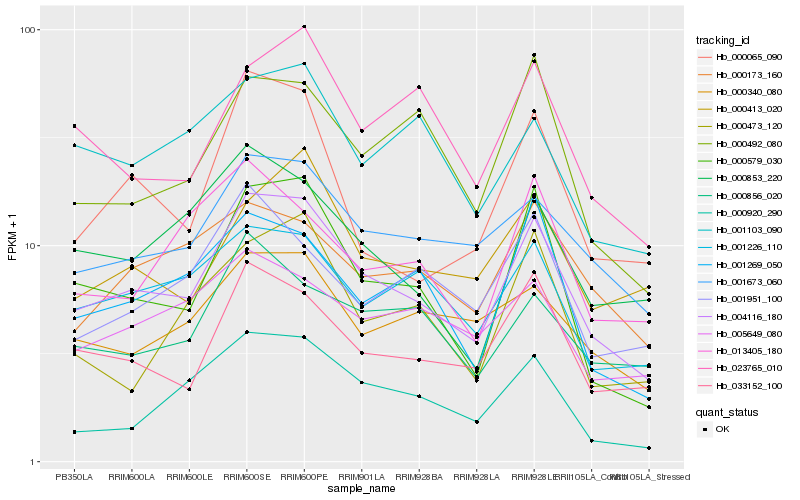
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004116_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646147 [Jatropha curcas] |
| 2 |
Hb_000579_030 |
0.1061523474 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 3 [Jatropha curcas] |
| 3 |
Hb_001673_060 |
0.1082347758 |
- |
- |
PREDICTED: uncharacterized protein LOC105635789 [Jatropha curcas] |
| 4 |
Hb_033152_100 |
0.1113244072 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_001226_110 |
0.1217717631 |
- |
- |
PREDICTED: uncharacterized protein LOC105638106 [Jatropha curcas] |
| 6 |
Hb_000920_290 |
0.1255287324 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 7 |
Hb_000340_080 |
0.1285463905 |
- |
- |
PREDICTED: isoamylase 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001269_050 |
0.1301451025 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |
| 9 |
Hb_000853_220 |
0.1307080002 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 10 |
Hb_000173_160 |
0.1332600924 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000856_020 |
0.1345443188 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 12 |
Hb_000492_080 |
0.1348614751 |
- |
- |
PREDICTED: probable sucrose-phosphate synthase 1 [Jatropha curcas] |
| 13 |
Hb_000413_020 |
0.1357292158 |
- |
- |
PREDICTED: U-box domain-containing protein 8 [Jatropha curcas] |
| 14 |
Hb_000065_090 |
0.1362333738 |
- |
- |
potassium channel tetramerization domain-containing protein, putative [Ricinus communis] |
| 15 |
Hb_023765_010 |
0.1366712202 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 16 |
Hb_005649_080 |
0.1369491685 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 17 |
Hb_001103_090 |
0.1387682244 |
- |
- |
PREDICTED: patellin-3 [Populus euphratica] |
| 18 |
Hb_000473_120 |
0.1432136242 |
- |
- |
hypothetical protein JCGZ_16177 [Jatropha curcas] |
| 19 |
Hb_013405_180 |
0.1436177321 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 20 |
Hb_001951_100 |
0.143737655 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |