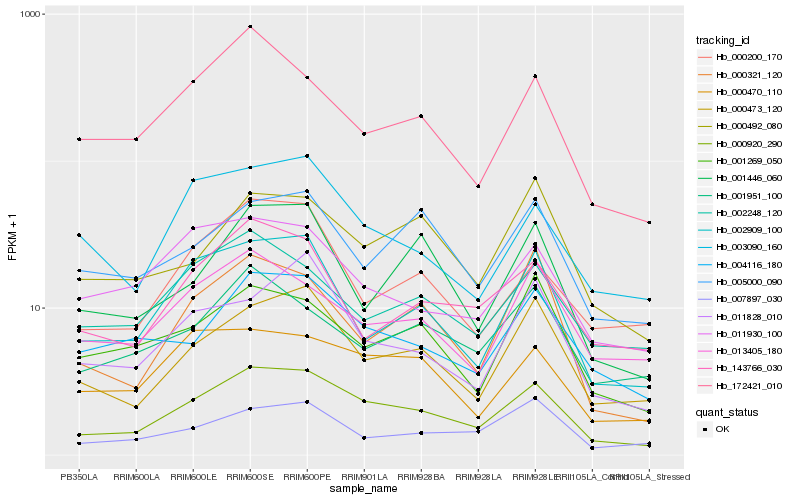
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000920_290 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 2 |
Hb_000473_120 |
0.116218879 |
- |
- |
hypothetical protein JCGZ_16177 [Jatropha curcas] |
| 3 |
Hb_004116_180 |
0.1255287324 |
- |
- |
PREDICTED: uncharacterized protein LOC105646147 [Jatropha curcas] |
| 4 |
Hb_002909_100 |
0.132341264 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 5 |
Hb_000470_110 |
0.1407930112 |
- |
- |
PREDICTED: probable LIM domain-containing serine/threonine-protein kinase DDB_G0287001 [Jatropha curcas] |
| 6 |
Hb_143766_030 |
0.14325257 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_000200_170 |
0.1445518241 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Populus euphratica] |
| 8 |
Hb_000492_080 |
0.1454449967 |
- |
- |
PREDICTED: probable sucrose-phosphate synthase 1 [Jatropha curcas] |
| 9 |
Hb_013405_180 |
0.1454496632 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 10 |
Hb_172421_010 |
0.1456242496 |
- |
- |
cysteine protease [Hevea brasiliensis] |
| 11 |
Hb_003090_160 |
0.1460516137 |
- |
- |
- |
| 12 |
Hb_002248_120 |
0.148276696 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 13 |
Hb_000321_120 |
0.1488376492 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 14 |
Hb_007897_030 |
0.1488495493 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR2 [Jatropha curcas] |
| 15 |
Hb_001951_100 |
0.1498193336 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 16 |
Hb_011828_010 |
0.1499490296 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 17 |
Hb_001446_060 |
0.1501216944 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 18 |
Hb_011930_100 |
0.1501445849 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 19 |
Hb_001269_050 |
0.1505252898 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |
| 20 |
Hb_005000_090 |
0.1505955021 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |