| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
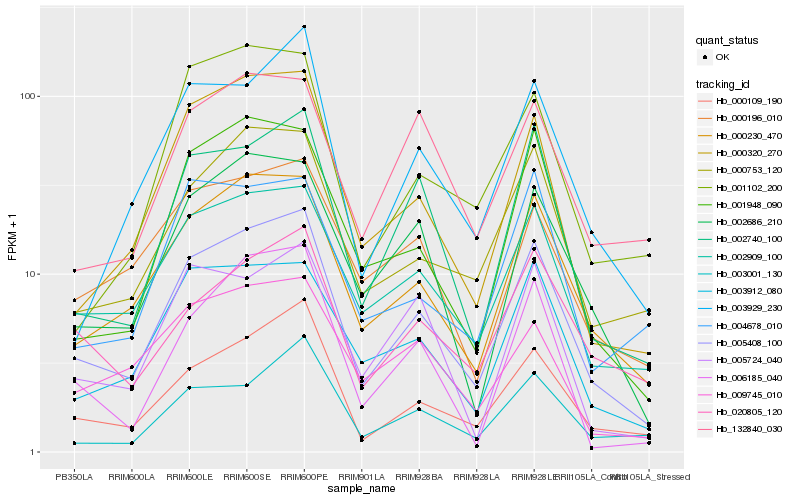
Hb_005408_100 |
0.0 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 2 |
Hb_000230_470 |
0.0838825252 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000109_190 |
0.0932427676 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 4 |
Hb_002909_100 |
0.0977469555 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 5 |
Hb_000320_270 |
0.1077592873 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002740_100 |
0.1099109034 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 7 |
Hb_000753_120 |
0.1115487411 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 8 |
Hb_003912_080 |
0.1192644973 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 9 |
Hb_002686_210 |
0.1199391369 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_001948_090 |
0.121934888 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0005s14540g [Populus trichocarpa] |
| 11 |
Hb_003929_230 |
0.1243815917 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 5 [Nelumbo nucifera] |
| 12 |
Hb_003001_130 |
0.1343464466 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006185_040 |
0.1366068874 |
- |
- |
PREDICTED: verprolin [Jatropha curcas] |
| 14 |
Hb_000196_010 |
0.1368826085 |
- |
- |
Patellin-5, putative [Ricinus communis] |
| 15 |
Hb_020805_120 |
0.1385199848 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 16 |
Hb_009745_010 |
0.1399086984 |
- |
- |
cellulose synthase-like protein [Populus tomentosa] |
| 17 |
Hb_005724_040 |
0.1415798201 |
- |
- |
hypothetical protein JCGZ_13142 [Jatropha curcas] |
| 18 |
Hb_001102_200 |
0.1440222493 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 19 |
Hb_004678_010 |
0.1456960523 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 20 |
Hb_132840_030 |
0.1487108131 |
transcription factor |
TF Family: MYB-related |
lhy1 [Populus tremula] |