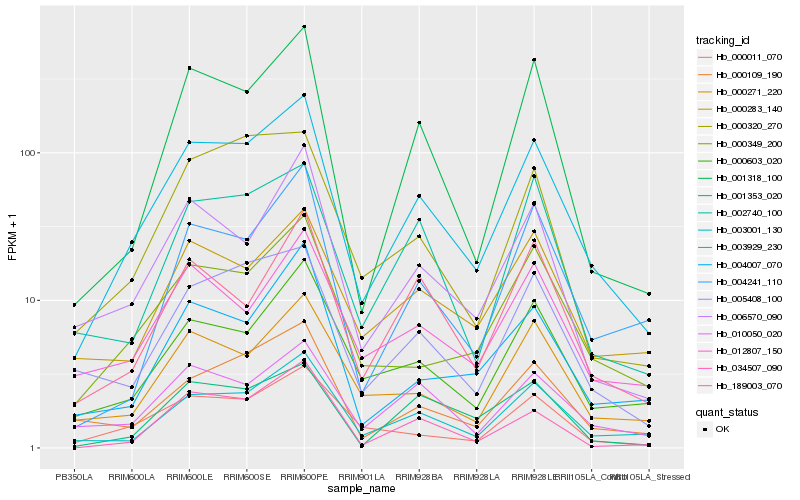
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003929_230 |
0.0 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 5 [Nelumbo nucifera] |
| 2 |
Hb_003001_130 |
0.1093581805 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001318_100 |
0.122603893 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 4 |
Hb_005408_100 |
0.1243815917 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 5 |
Hb_000349_200 |
0.1252564184 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g12500 [Jatropha curcas] |
| 6 |
Hb_000109_190 |
0.1278715491 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 7 |
Hb_000603_020 |
0.131692806 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 8 |
Hb_010050_020 |
0.1362973796 |
- |
- |
PREDICTED: uncharacterized protein LOC102601222 [Solanum tuberosum] |
| 9 |
Hb_006570_090 |
0.1364231674 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 10 |
Hb_002740_100 |
0.1379489363 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 11 |
Hb_000011_070 |
0.1383995402 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 12 |
Hb_000271_220 |
0.1412818975 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 13 |
Hb_034507_090 |
0.1426798626 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 14 |
Hb_004241_110 |
0.1431592099 |
- |
- |
Minor allergen Alt a, putative [Ricinus communis] |
| 15 |
Hb_000320_270 |
0.1433013418 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004007_070 |
0.1447027244 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 17 |
Hb_000283_140 |
0.1451786778 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001353_020 |
0.1458138357 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_189003_070 |
0.1467302205 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 20 |
Hb_012807_150 |
0.1487487433 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |