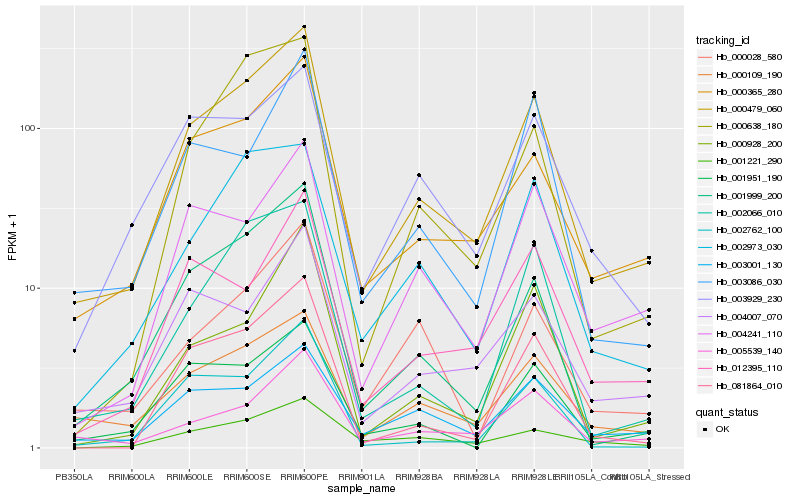
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000479_060 |
0.0 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1 [Vitis vinifera] |
| 2 |
Hb_000365_280 |
0.094774706 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 3 |
Hb_000109_190 |
0.1167228905 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 4 |
Hb_001999_200 |
0.1197483645 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005539_140 |
0.1328762951 |
- |
- |
PREDICTED: U-box domain-containing protein 19-like [Jatropha curcas] |
| 6 |
Hb_002973_030 |
0.1380891219 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 7 |
Hb_081864_010 |
0.1386837611 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_004007_070 |
0.1391497324 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 9 |
Hb_003001_130 |
0.1392680726 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002066_010 |
0.1431756671 |
- |
- |
PREDICTED: acetylglutamate kinase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001951_190 |
0.1442167107 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 3-like isoform X2 [Populus euphratica] |
| 12 |
Hb_000638_180 |
0.1452404136 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 13 |
Hb_004241_110 |
0.1463900643 |
- |
- |
Minor allergen Alt a, putative [Ricinus communis] |
| 14 |
Hb_012395_110 |
0.1513554571 |
- |
- |
PREDICTED: peroxygenase [Jatropha curcas] |
| 15 |
Hb_003929_230 |
0.15150751 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 5 [Nelumbo nucifera] |
| 16 |
Hb_000028_580 |
0.1519653308 |
- |
- |
PREDICTED: glutamate decarboxylase 5-like [Jatropha curcas] |
| 17 |
Hb_001221_290 |
0.1535007771 |
- |
- |
hypothetical protein RCOM_1506010 [Ricinus communis] |
| 18 |
Hb_002762_100 |
0.1548559201 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_000928_200 |
0.1553290197 |
- |
- |
PREDICTED: RING-H2 finger protein ATL52-like [Jatropha curcas] |
| 20 |
Hb_003086_030 |
0.1558593658 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |