| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
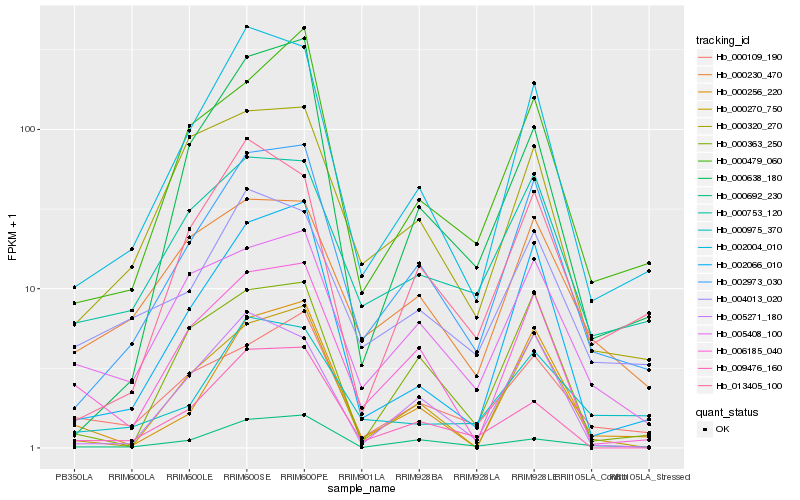
Hb_002973_030 |
0.0 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 2 |
Hb_002004_010 |
0.1028472351 |
- |
- |
CBL-interacting protein kinase 9 [Populus trichocarpa] |
| 3 |
Hb_002066_010 |
0.1308047727 |
- |
- |
PREDICTED: acetylglutamate kinase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000479_060 |
0.1380891219 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1 [Vitis vinifera] |
| 5 |
Hb_000753_120 |
0.1435835265 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 6 |
Hb_000270_750 |
0.1438455893 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46-like [Populus euphratica] |
| 7 |
Hb_013405_100 |
0.1495841753 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 8 [Jatropha curcas] |
| 8 |
Hb_000363_250 |
0.1507244214 |
- |
- |
PREDICTED: pirin-like protein [Malus domestica] |
| 9 |
Hb_006185_040 |
0.1514845388 |
- |
- |
PREDICTED: verprolin [Jatropha curcas] |
| 10 |
Hb_000692_230 |
0.152805083 |
- |
- |
phospholipase d delta, putative [Ricinus communis] |
| 11 |
Hb_000320_270 |
0.1532158902 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_005408_100 |
0.1535994973 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 13 |
Hb_000109_190 |
0.153952521 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 14 |
Hb_004013_020 |
0.1542345216 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |
| 15 |
Hb_000638_180 |
0.1563677759 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 16 |
Hb_005271_180 |
0.1578093035 |
- |
- |
Leucine-rich repeat receptor protein kinase EXS precursor, putative [Ricinus communis] |
| 17 |
Hb_000230_470 |
0.1583367006 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000256_220 |
0.1606213364 |
- |
- |
PREDICTED: flotillin-like protein 4 [Jatropha curcas] |
| 19 |
Hb_000975_370 |
0.1613116551 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB59 isoform X1 [Jatropha curcas] |
| 20 |
Hb_009476_160 |
0.1615082483 |
- |
- |
RING-H2 finger protein ATL2N, putative [Ricinus communis] |