| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
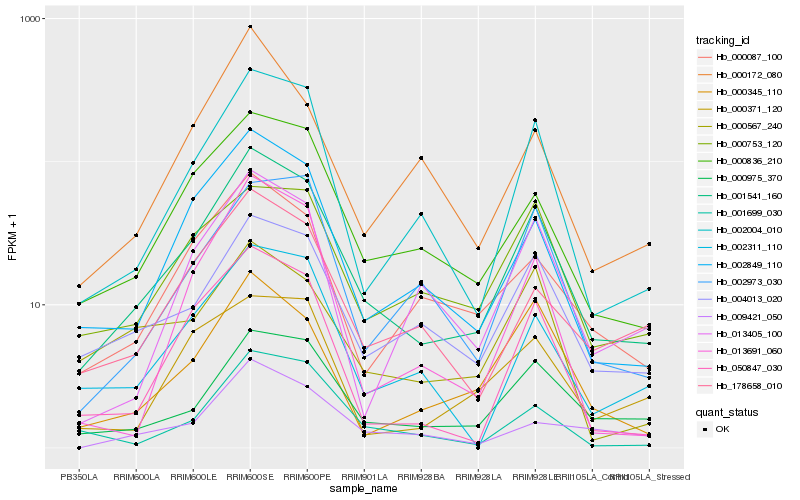
Hb_001541_160 |
0.0 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 2 |
Hb_002004_010 |
0.1242583976 |
- |
- |
CBL-interacting protein kinase 9 [Populus trichocarpa] |
| 3 |
Hb_000836_210 |
0.1246047014 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 4 |
Hb_002849_110 |
0.1296610249 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 5 |
Hb_000975_370 |
0.1310698899 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB59 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004013_020 |
0.1337613704 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |
| 7 |
Hb_050847_030 |
0.1498231095 |
- |
- |
PREDICTED: cytochrome P450 714A1-like [Populus euphratica] |
| 8 |
Hb_178658_010 |
0.1516393581 |
- |
- |
PREDICTED: uncharacterized protein LOC105641396 [Jatropha curcas] |
| 9 |
Hb_000087_100 |
0.1525703301 |
- |
- |
PREDICTED: cellulose synthase-like protein D3 [Jatropha curcas] |
| 10 |
Hb_002311_110 |
0.1533654565 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000345_110 |
0.1575222646 |
- |
- |
PREDICTED: uncharacterized protein LOC105647507 [Jatropha curcas] |
| 12 |
Hb_000753_120 |
0.1582667525 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 13 |
Hb_002973_030 |
0.1703527267 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 14 |
Hb_013405_100 |
0.1726140353 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 8 [Jatropha curcas] |
| 15 |
Hb_001699_030 |
0.1734036361 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_009421_050 |
0.1736897769 |
- |
- |
PREDICTED: uncharacterized protein LOC105647037 [Jatropha curcas] |
| 17 |
Hb_000371_120 |
0.1758826699 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_000172_080 |
0.1788440321 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 19 |
Hb_000567_240 |
0.1796994261 |
- |
- |
PREDICTED: F-box/LRR-repeat MAX2 homolog A-like [Jatropha curcas] |
| 20 |
Hb_013691_060 |
0.1797487322 |
- |
- |
PREDICTED: uncharacterized protein LOC105119066 [Populus euphratica] |