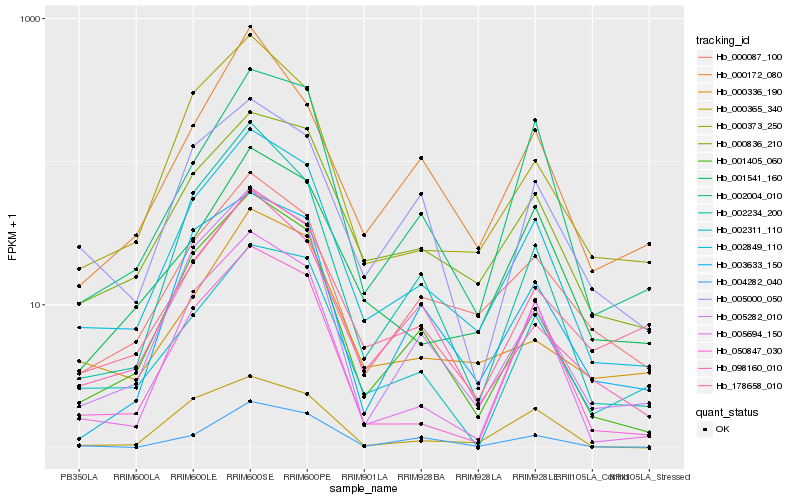
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002849_110 |
0.0 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 2 |
Hb_000836_210 |
0.0812270331 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 3 |
Hb_001405_060 |
0.0948296412 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 4-like [Jatropha curcas] |
| 4 |
Hb_005282_010 |
0.1070840168 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 12 [Populus euphratica] |
| 5 |
Hb_000373_250 |
0.11055468 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 6 |
Hb_000087_100 |
0.1140533892 |
- |
- |
PREDICTED: cellulose synthase-like protein D3 [Jatropha curcas] |
| 7 |
Hb_004282_040 |
0.1221898595 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_098160_010 |
0.1239506268 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_002234_200 |
0.124239124 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 10 |
Hb_005694_150 |
0.1254165547 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 11 |
Hb_178658_010 |
0.1268822162 |
- |
- |
PREDICTED: uncharacterized protein LOC105641396 [Jatropha curcas] |
| 12 |
Hb_002004_010 |
0.1284954947 |
- |
- |
CBL-interacting protein kinase 9 [Populus trichocarpa] |
| 13 |
Hb_001541_160 |
0.1296610249 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 14 |
Hb_002311_110 |
0.1319834408 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_050847_030 |
0.1322813589 |
- |
- |
PREDICTED: cytochrome P450 714A1-like [Populus euphratica] |
| 16 |
Hb_005000_050 |
0.1343146971 |
- |
- |
PREDICTED: tetraspanin-2 [Jatropha curcas] |
| 17 |
Hb_003633_150 |
0.1343623606 |
- |
- |
hypothetical protein JCGZ_23296 [Jatropha curcas] |
| 18 |
Hb_000172_080 |
0.1346909931 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 19 |
Hb_000365_340 |
0.1362624808 |
- |
- |
PREDICTED: uncharacterized protein LOC105649067 [Jatropha curcas] |
| 20 |
Hb_000336_190 |
0.1393625353 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Gossypium raimondii] |