| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
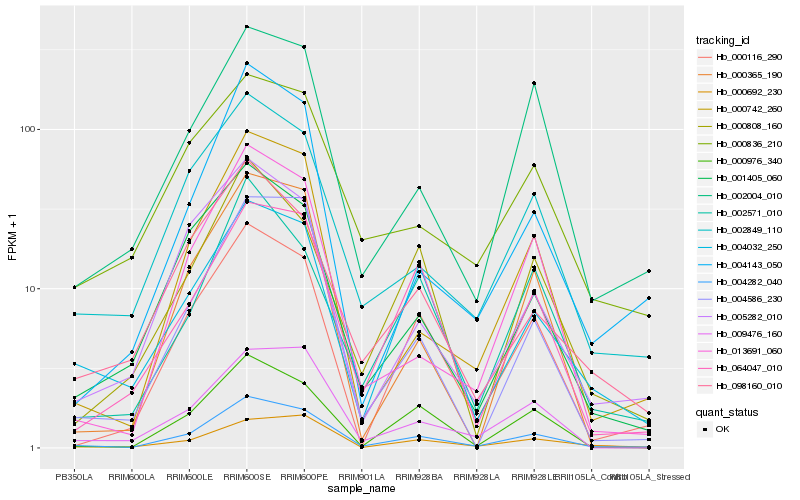
Hb_004282_040 |
0.0 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_000116_290 |
0.1214428491 |
- |
- |
PREDICTED: proton-coupled amino acid transporter 3 [Jatropha curcas] |
| 3 |
Hb_002849_110 |
0.1221898595 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 4 |
Hb_000742_260 |
0.1281222922 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |
| 5 |
Hb_009476_160 |
0.1317467577 |
- |
- |
RING-H2 finger protein ATL2N, putative [Ricinus communis] |
| 6 |
Hb_000976_340 |
0.1318037563 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_000692_230 |
0.1328276767 |
- |
- |
phospholipase d delta, putative [Ricinus communis] |
| 8 |
Hb_004032_250 |
0.1333461136 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002004_010 |
0.1373826467 |
- |
- |
CBL-interacting protein kinase 9 [Populus trichocarpa] |
| 10 |
Hb_064047_010 |
0.1390326846 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001405_060 |
0.1403152999 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 4-like [Jatropha curcas] |
| 12 |
Hb_004586_230 |
0.1423373447 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_013691_060 |
0.145546881 |
- |
- |
PREDICTED: uncharacterized protein LOC105119066 [Populus euphratica] |
| 14 |
Hb_005282_010 |
0.1480350578 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 12 [Populus euphratica] |
| 15 |
Hb_098160_010 |
0.1496503122 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_000836_210 |
0.150499643 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 17 |
Hb_002571_010 |
0.1507179766 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 18 |
Hb_004143_050 |
0.1539898828 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |
| 19 |
Hb_000808_160 |
0.1549826645 |
- |
- |
PREDICTED: uncharacterized protein LOC105647443 [Jatropha curcas] |
| 20 |
Hb_000365_190 |
0.1551874049 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |