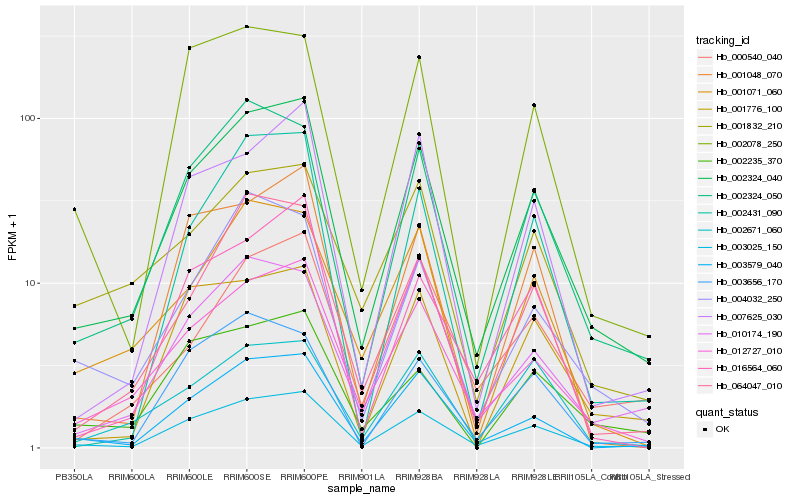
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002324_040 |
0.0 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 2 |
Hb_003025_150 |
0.0569056965 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_012727_010 |
0.0803141856 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 4 |
Hb_002324_050 |
0.1013814817 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 5 |
Hb_003579_040 |
0.1049305887 |
- |
- |
PREDICTED: monoacylglycerol lipase ABHD6 [Jatropha curcas] |
| 6 |
Hb_064047_010 |
0.1176494957 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004032_250 |
0.1214013917 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001071_060 |
0.136663666 |
- |
- |
Trehalose phosphatase/synthase 5 isoform 1 [Theobroma cacao] |
| 9 |
Hb_007625_030 |
0.1372738147 |
- |
- |
unknown [Medicago truncatula] |
| 10 |
Hb_000540_040 |
0.1396791577 |
- |
- |
PREDICTED: chitinase 2 [Amborella trichopoda] |
| 11 |
Hb_002431_090 |
0.1400345181 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003656_170 |
0.1406707346 |
- |
- |
PREDICTED: phenylalanine ammonia-lyase-like [Jatropha curcas] |
| 13 |
Hb_002235_370 |
0.1436353477 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 14 |
Hb_002671_060 |
0.1449818781 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 15 |
Hb_001832_210 |
0.1451632535 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 16 |
Hb_016564_060 |
0.1451715925 |
- |
- |
PREDICTED: uncharacterized protein LOC105628249 [Jatropha curcas] |
| 17 |
Hb_002078_250 |
0.1451803468 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001048_070 |
0.1456991434 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 19 |
Hb_010174_190 |
0.1464779637 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 20 |
Hb_001776_100 |
0.1465859816 |
- |
- |
PREDICTED: outer envelope pore protein 16, chloroplastic [Jatropha curcas] |