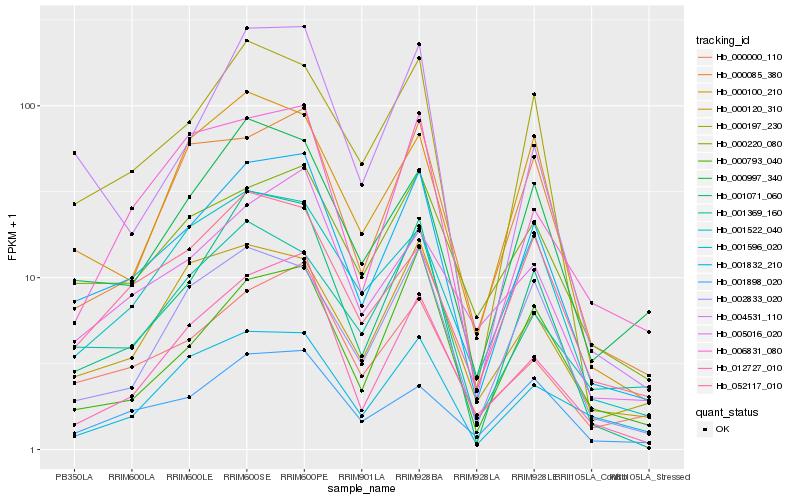
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001832_210 |
0.0 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 2 |
Hb_001071_060 |
0.0992984719 |
- |
- |
Trehalose phosphatase/synthase 5 isoform 1 [Theobroma cacao] |
| 3 |
Hb_000000_110 |
0.1105332063 |
- |
- |
hypothetical protein CISIN_1g0027772mg, partial [Citrus sinensis] |
| 4 |
Hb_000197_230 |
0.1110326588 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 5 |
Hb_000997_340 |
0.1172505801 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 6 |
Hb_000220_080 |
0.1188673482 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_000085_380 |
0.1253146353 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001898_020 |
0.1274464531 |
- |
- |
PREDICTED: uncharacterized protein LOC105629659 [Jatropha curcas] |
| 9 |
Hb_012727_010 |
0.1279760226 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 10 |
Hb_001522_040 |
0.1314247187 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_000120_310 |
0.1316892303 |
- |
- |
Gibberellin 3-beta-dioxygenase, putative [Ricinus communis] |
| 12 |
Hb_002833_020 |
0.1322123408 |
desease resistance |
Gene Name: LRR_4 |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 13 |
Hb_005016_020 |
0.1343790315 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_004531_110 |
0.1349500944 |
- |
- |
PREDICTED: aquaporin TIP2-1 [Jatropha curcas] |
| 15 |
Hb_052117_010 |
0.1351870766 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_001596_020 |
0.135526564 |
- |
- |
hypothetical protein CICLE_v10014140mg [Citrus clementina] |
| 17 |
Hb_001369_160 |
0.1364574862 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 18 |
Hb_006831_080 |
0.1371615498 |
- |
- |
PREDICTED: uncharacterized protein LOC105642126 [Jatropha curcas] |
| 19 |
Hb_000793_040 |
0.1375392252 |
- |
- |
PREDICTED: transcription factor LHW [Jatropha curcas] |
| 20 |
Hb_000100_210 |
0.1384155004 |
- |
- |
PREDICTED: selenium-binding protein 1-like [Jatropha curcas] |