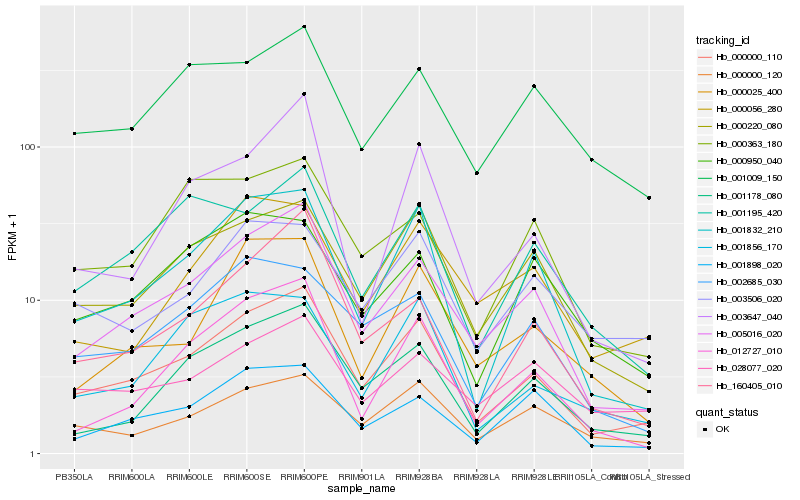
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000000_110 |
0.0 |
- |
- |
hypothetical protein CISIN_1g0027772mg, partial [Citrus sinensis] |
| 2 |
Hb_005016_020 |
0.0863688773 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_001832_210 |
0.1105332063 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 4 |
Hb_000220_080 |
0.1159693482 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_001178_080 |
0.12110708 |
- |
- |
PREDICTED: protein kinase PVPK-1 [Jatropha curcas] |
| 6 |
Hb_000000_120 |
0.1304337827 |
- |
- |
hypothetical protein EUGRSUZ_A004731, partial [Eucalyptus grandis] |
| 7 |
Hb_002685_030 |
0.1366459402 |
- |
- |
PREDICTED: protein SMG7L [Jatropha curcas] |
| 8 |
Hb_160405_010 |
0.1381446383 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 9 |
Hb_003506_020 |
0.1439767389 |
- |
- |
PREDICTED: uncharacterized protein LOC105125881 isoform X1 [Populus euphratica] |
| 10 |
Hb_001195_420 |
0.1445643302 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X4 [Jatropha curcas] |
| 11 |
Hb_001898_020 |
0.1446141318 |
- |
- |
PREDICTED: uncharacterized protein LOC105629659 [Jatropha curcas] |
| 12 |
Hb_012727_010 |
0.1450903349 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 13 |
Hb_000025_400 |
0.1453326049 |
- |
- |
PREDICTED: uncharacterized protein LOC105110770 [Populus euphratica] |
| 14 |
Hb_001856_170 |
0.1468679613 |
- |
- |
PREDICTED: ABC transporter B family member 27 [Jatropha curcas] |
| 15 |
Hb_001009_150 |
0.1473925245 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase [Gossypium arboreum] |
| 16 |
Hb_028077_020 |
0.1514256997 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000363_180 |
0.1525351358 |
- |
- |
PREDICTED: uncharacterized protein LOC105633052 [Jatropha curcas] |
| 18 |
Hb_003647_040 |
0.1538637982 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000056_280 |
0.1546223367 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 20 |
Hb_000950_040 |
0.1550488243 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |