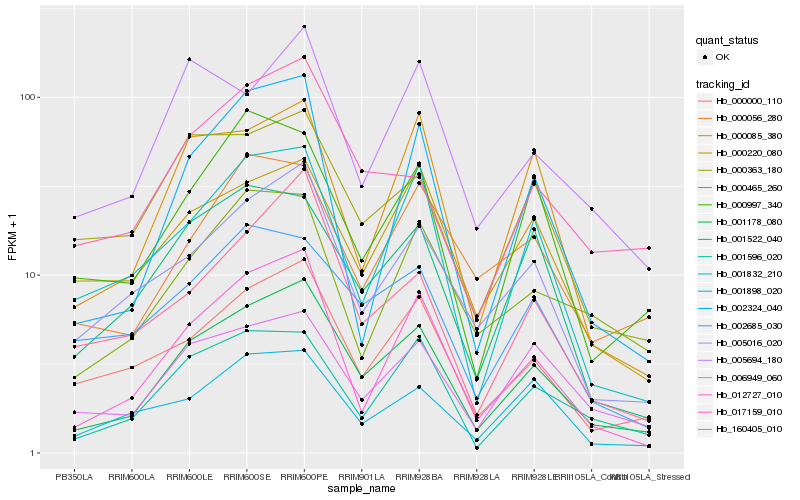
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001178_080 |
0.0 |
- |
- |
PREDICTED: protein kinase PVPK-1 [Jatropha curcas] |
| 2 |
Hb_005016_020 |
0.1192635118 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_012727_010 |
0.1200004928 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 4 |
Hb_000000_110 |
0.12110708 |
- |
- |
hypothetical protein CISIN_1g0027772mg, partial [Citrus sinensis] |
| 5 |
Hb_001596_020 |
0.1301271514 |
- |
- |
hypothetical protein CICLE_v10014140mg [Citrus clementina] |
| 6 |
Hb_006949_060 |
0.1347895178 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 7 |
Hb_017159_010 |
0.1394708162 |
- |
- |
- |
| 8 |
Hb_001832_210 |
0.1415250949 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 9 |
Hb_001522_040 |
0.1431590835 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_000085_380 |
0.1462404727 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_160405_010 |
0.1463970229 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 12 |
Hb_000363_180 |
0.1468998687 |
- |
- |
PREDICTED: uncharacterized protein LOC105633052 [Jatropha curcas] |
| 13 |
Hb_000220_080 |
0.1482268471 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_002685_030 |
0.1495855495 |
- |
- |
PREDICTED: protein SMG7L [Jatropha curcas] |
| 15 |
Hb_001898_020 |
0.1499029874 |
- |
- |
PREDICTED: uncharacterized protein LOC105629659 [Jatropha curcas] |
| 16 |
Hb_000997_340 |
0.1502593883 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 17 |
Hb_002324_040 |
0.1505480628 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 18 |
Hb_005694_180 |
0.1510578222 |
- |
- |
PREDICTED: cytochrome b5 [Jatropha curcas] |
| 19 |
Hb_000465_260 |
0.1532103696 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT4-like [Jatropha curcas] |
| 20 |
Hb_000056_280 |
0.153409248 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |