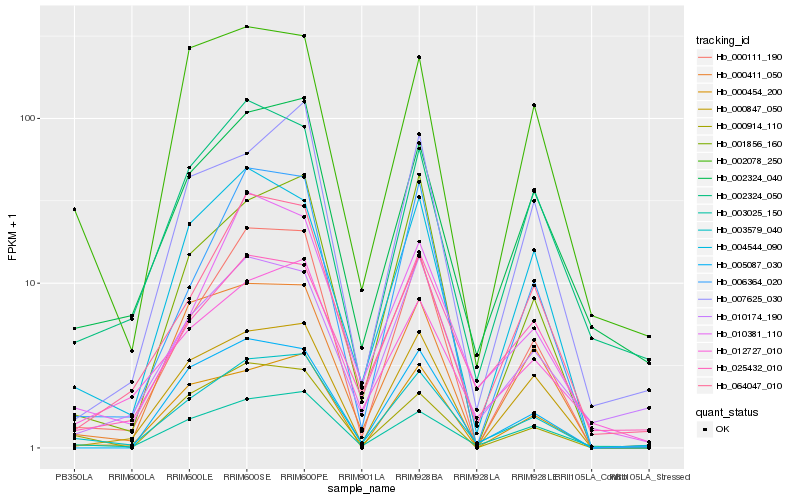
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003579_040 |
0.0 |
- |
- |
PREDICTED: monoacylglycerol lipase ABHD6 [Jatropha curcas] |
| 2 |
Hb_003025_150 |
0.0914579835 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002324_040 |
0.1049305887 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 4 |
Hb_000111_190 |
0.1098598406 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1 [Jatropha curcas] |
| 5 |
Hb_012727_010 |
0.1299290246 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 6 |
Hb_000914_110 |
0.1320426755 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein WIP2 [Jatropha curcas] |
| 7 |
Hb_004544_090 |
0.1341416285 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 8 |
Hb_000411_050 |
0.1354718836 |
- |
- |
hypothetical protein AMTR_s00066p00166230 [Amborella trichopoda] |
| 9 |
Hb_000454_200 |
0.1357205857 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG7 [Jatropha curcas] |
| 10 |
Hb_000847_050 |
0.1367749413 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 11 |
Hb_010174_190 |
0.13700751 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 12 |
Hb_002324_050 |
0.1381070315 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 13 |
Hb_001856_160 |
0.1381749282 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006364_020 |
0.1398634588 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM1 [Jatropha curcas] |
| 15 |
Hb_010381_110 |
0.1408690432 |
- |
- |
PRA1 family protein [Theobroma cacao] |
| 16 |
Hb_025432_010 |
0.1423774051 |
- |
- |
hypothetical protein JCGZ_21627 [Jatropha curcas] |
| 17 |
Hb_002078_250 |
0.1423923714 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007625_030 |
0.14264472 |
- |
- |
unknown [Medicago truncatula] |
| 19 |
Hb_005087_030 |
0.1438198035 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 20 |
Hb_064047_010 |
0.1438466451 |
- |
- |
conserved hypothetical protein [Ricinus communis] |